GLBRC Data Sets
Highlighted below are a variety of published studies that include data sets that might be of interest to the scientific community and have been deposited in online data repositories. Only data sets published in GLBRC-approved repositories following the FAIR Guiding Principles are highlighted. More information can be found on our guidelines page.
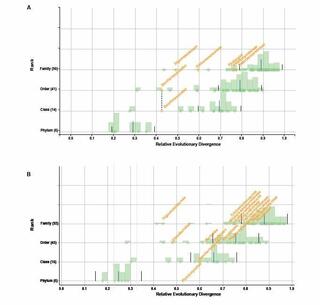
A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina
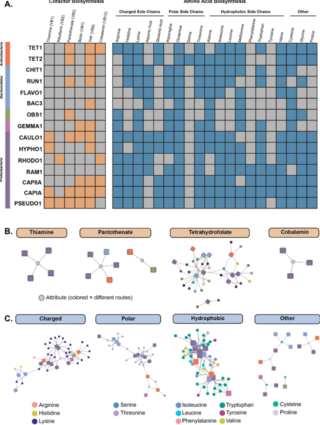
The subphylum Saccharomycotina is a lineage in the fungal phylum Ascomycota that exhibits levels of genomic diversity similar to those of plants and animals. The Saccharomycotina consist of more than 1 200 known species currently divided into 16 families, one order, and one class.
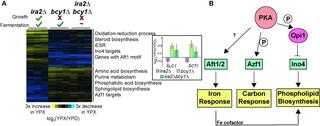
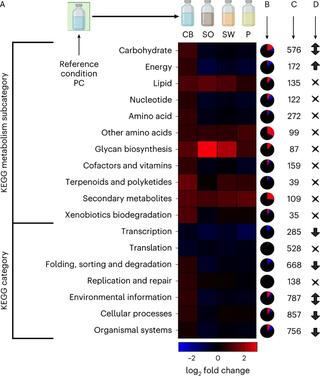
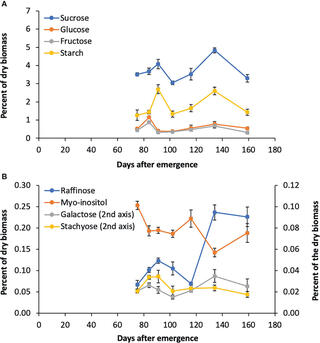
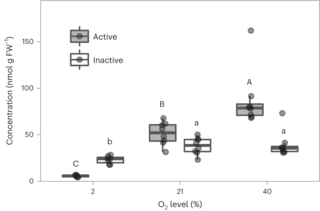
PKA regulatory subunit Bcy1 couples growth, lipid metabolism, and fermentation during anaerobic xylose growth in Saccharomyces cerevisiae
Organisms have evolved elaborate physiological pathways that regulate growth, proliferation, metabolism, and stress response. These pathways must be properly coordinated to elicit the appropriate response to an ever-changing environment.
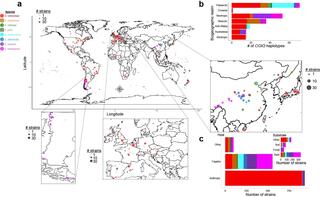
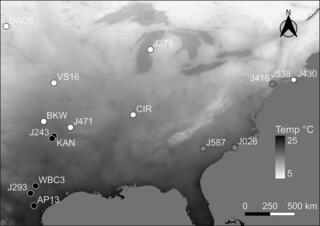
Macroevolutionary diversity of traits and genomes in the model yeast genus Saccharomyces
Species is the fundamental unit to quantify biodiversity. In recent years, the model yeast Saccharomyces cerevisiae has seen an increased number of studies related to its geographical distribution, population structure, and phenotypic diversity.
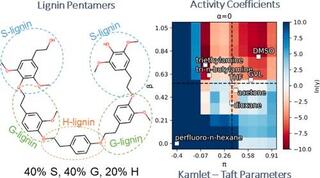
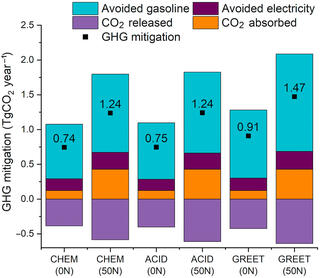
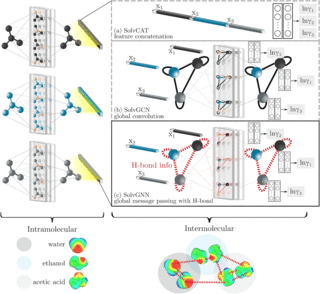
Heuristic computational model for predicting lignin solubility in tailored organic solvents
Lignin is a random heteropolymer that has been extensively studied as a renewable source of aromatic precursors for high-value chemicals, biofuels, and bioplastics. A key challenge in lignin valorization is the structural and compositional heterogeneity of lignin feedstocks.
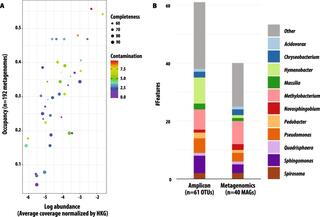
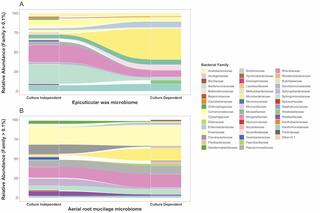
Seasonal activities of the phyllosphere microbiome of perennial crops
Understanding the interactions between plants and microorganisms can inform microbiome management to enhance crop productivity and resilience to stress. Here, we apply a genome-centric approach to identify ecologically important leaf microbiome members on replicated plots of field-grown switchgrass and miscanthus, and to quantify their activities over two growing seasons for switchgrass.
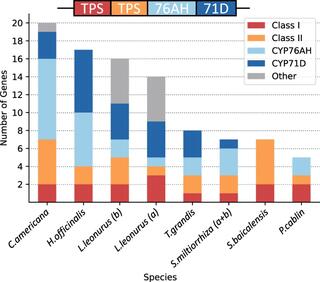
Uncovering a miltiradiene biosynthetic gene cluster in the Lamiaceae reveals a dynamic evolutionary trajectory
The spatial organization of genes within plant genomes can drive evolution of specialized metabolic pathways. Terpenoids are important specialized metabolites in plants with diverse adaptive functions that enable environmental interactions. Here, we report the genome assemblies of Prunella vulgaris, Plectranthus barbatus, and Leonotis leonurus.
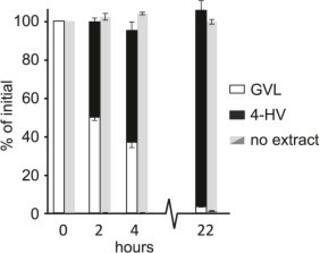
Lignin deconstruction by anaerobic fungi
Lignocellulose forms plant cell walls, and its three constituent polymers, cellulose, hemicellulose and lignin, represent the largest renewable organic carbon pool in the terrestrial biosphere.
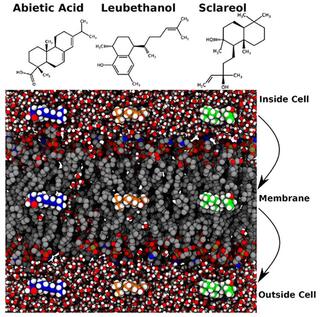
Plant terpenoid permeability through biological membranes explored via molecular simulations
Plants synthesize small molecule diterpenes composed of 20 carbons from precursor isopentenyl diphosphate and dimethylallyl disphosphate, manufacturing diverse compounds used for defense, signaling, and other functions. Industrially, diterpenes are used as natural aromas and flavoring, as pharmaceuticals, and as natural insecticides or repellents.
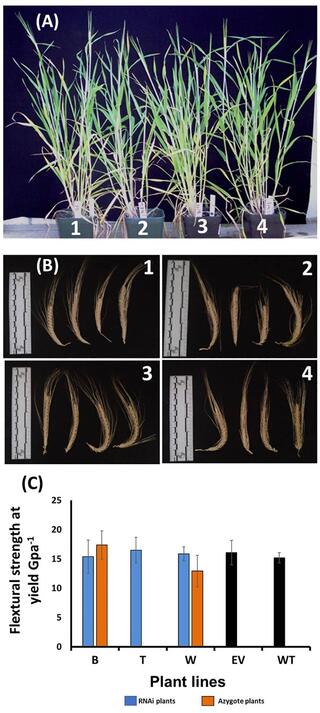
Downregulation of barley ferulate 5-hydroxylase dramatically alters straw lignin structure without impact on mechanical properties
Barley is a major cereal crop for temperate climates, and a diploid genetic model for polyploid wheat. Cereal straw biomass is an attractive source of feedstock for green technologies but lignin, a key determinant of feedstock recalcitrance, complicates bio-conversion processes. However, manipulating lignin content to improve the conversion process could negatively affect agronomic traits.
Pest suppression potential varies across 10 bioenergy cropping systems
Top-down suppression of herbivores is a fundamental ecological process and a critical service in agricultural landscapes. Adoption of bioenergy cropping systems is likely to become an increasingly important driver causing loss or gain of this service in coming decades.