Seasonal activities of the phyllosphere microbiome of perennial crops
A. Howe et al. "Seasonal activities of the phyllosphere microbiome of perennial crops" Nature Communications 14:1039 (2023) [DOI:10.1038/s41467-023-36515-y]
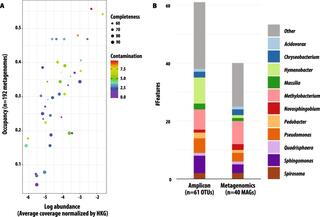
Understanding the interactions between plants and microorganisms can inform microbiome management to enhance crop productivity and resilience to stress. Here, we apply a genome-centric approach to identify ecologically important leaf microbiome members on replicated plots of field-grown switchgrass and miscanthus, and to quantify their activities over two growing seasons for switchgrass. We use metagenome and metatranscriptome sequencing and curate 40 medium- and high-quality metagenome-assembled-genomes (MAGs). We find that classes represented by these MAGs (Actinomycetia, Alpha- and Gamma- Proteobacteria, and Bacteroidota) are active in the late season, and upregulate transcripts for short-chain dehydrogenase, molybdopterin oxidoreductase, and polyketide cyclase. Stress-associated pathways are expressed for most MAGs, suggesting engagement with the host environment. We also detect seasonally activated biosynthetic pathways for terpenes and various non-ribosomal peptide pathways that are poorly annotated. Our findings support that leaf-associated bacterial populations are seasonally dynamic and responsive to host cues.
The raw and processed metagenome and metatranscriptome data generated in this study have been deposited in the Joint Genome Institute Genome Portal database under proposal ID 503249 [https://genome.jgi.doe.gov/portal/Seadynanfunction/Seadynanfunction.inf…]. The MAGs data generated in this study have been depositied in NCBI under bioproject PRJNA800073. The metadata, including metadata standards for metagenomes, metatranscriptomes, and metagenome-assembled genomes, for this study are provided in the Supplementary Data. Source data are provided with this paper.
Annotated code and links to data and metadata are available on GitHub (https://github.com/ShadeLab/PAPER_Howe_2021_switchgrass_MetaT) and Zenodo (https://zenodo.org/record/10040).