Macroevolutionary diversity of traits and genomes in the model yeast genus Saccharomyces
D. Peris et al. "Macroevolutionary diversity of traits and genomes in the model yeast genus Saccharomyces" Nature Communications 14:690 (2023) [DOI:10.1038/s41467-023-36139-2]
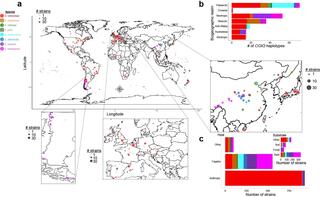
Species is the fundamental unit to quantify biodiversity. In recent years, the model yeast Saccharomyces cerevisiae has seen an increased number of studies related to its geographical distribution, population structure, and phenotypic diversity. However, seven additional species from the same genus have been less thoroughly studied, which has limited our understanding of the macroevolutionary events leading to the diversification of this genus over the last 20 million years. Here, we show the geographies, hosts, substrates, and phylogenetic relationships for approximately 1,800 Saccharomyces strains, covering the complete genus with unprecedented breadth and depth. We generated and analyzed complete genome sequences of 163 strains and phenotyped 128 phylogenetically diverse strains. This dataset provides insights about genetic and phenotypic diversity within and between species and populations, quantifies reticulation and incomplete lineage sorting, and demonstrates how gene flow and selection have affected traits, such as galactose metabolism. These findings elevate the genus Saccharomyces as a model to understand biodiversity and evolution in microbial eukaryotes.
The COX2 and COX3 sequences generated in this study were deposited in GenBank under accession nos. MH813536-MH813939. The GAL genes that were Sanger-sequenced in this study were deposited in GenBank under accession nos. OL660614-OL660618. Illumina sequencing data generated in this study have been deposited in NCBI’s SRA database under accession Bioproject code PRJNA475869. Genome assemblies and annotations generated in this study are available on the European Nucleotide Archive (ENA) under project accession code PRJEB48264. Accession numbers of downloaded Illumina sequences or genome assemblies are provided in Supplementary Data. Details regarding the location of source data for Figs. 2–6, as well as Supplementary Figs. 3–15, and 17–29 can be found under the “Source Data” heading of the Github repository. Raw data generated in this study is deposited in FigShare.