GLBRC Data Sets
Highlighted below are a variety of published studies that include data sets that might be of interest to the scientific community and have been deposited in online data repositories. Only data sets published in GLBRC-approved repositories following the FAIR Guiding Principles are highlighted. More information can be found on our guidelines page.
GLBRC Sustainability Data Catalog
This Data Catalog is a collection of data from GLBRC's Sustainability research carried out in Michigan and Wisconsin. The Data Catalog summarizes each data table in order to allow the GLBRC community to better understand the data that have been collected and encourage collaboration.
Transcriptomic data sets examining several stress responses in Zymomonas mobilis strains
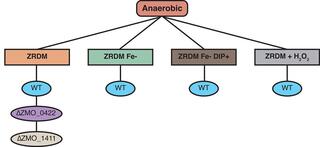
We used RNA-seq to compare gene expression from Zymomonas mobilis ZM4 grown under various conditions: aerobic ± paraquat, anaerobic ± hydrogen peroxide, or an iron chelator. We analyzed two mutant strains lacking predicted transcription factors (ZMO_0442 and ZMO_1411) grown under aerobic or anaerobic conditions. We report the RNA-seq data from these experiments.
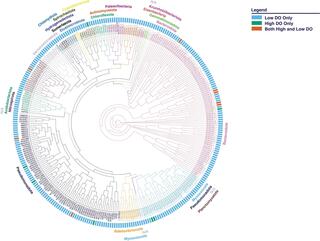
Metagenomes and metagenome-assembled genomes from a nutrient removal plant at Los Angeles County Sanitation Districts (LACSD) that transitioned from high to low dissolved oxygen
Operating biological nutrient removal (BNR) wastewater treatment plants with low dissolved oxygen (DO) conditions can reduce energy costs. We report on five metagenomes and 492 metagenome-assembled genomes (MAGs) obtained from samples collected at the Pomona water reclamation plant before and after a DO reduction from 3.5 to 0.7 mg/L.
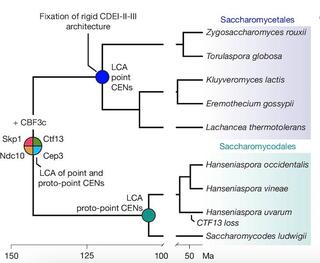
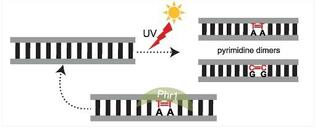
Ancient co-option of LTR retrotransposons as yeast centromeres
Centromeres ensure accurate chromosome segregation, yet their DNA evolves rapidly across eukaryotes leaving the origins of new centromere architectures unclear1,2,3,4. The brewer’s yeast Saccharomyces cerevisiae exemplifies this long-standing puzzle.
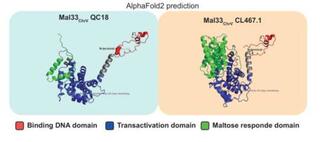
MAL33 drives natural variation in maltose metabolism in Saccharomyces eubayanus
Maltose is one of the most abundant sugars in brewer’s wort, and its efficient utilization is critical for successful fermentation. However, maltose consumption varies naturally among Saccharomyces eubayanus strains isolated from different host trees, such as Quercus and Nothofagus.
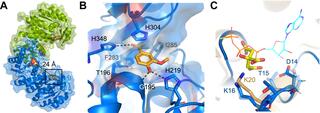
Energetic and structural control of polyspecificity in a multidrug transporter
Multidrug efflux pumps are dynamic molecular machines that drive antibiotic resistance by harnessing ion gradients to export chemically diverse substrates. Despite their clinical importance, the molecular principles underlying multidrug promiscuity and energy efficiency remain poorly understood.
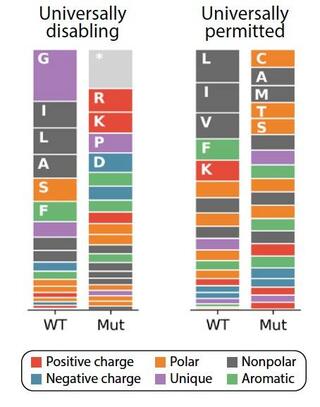
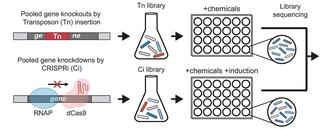
Orthogonal chemical genomics approaches reveal genomic targets for increasing anaerobic chemical tolerance in Zymomonas mobilis
Genetically engineered microbes have the potential to increase efficiency in the bioeconomy by overcoming growth-limiting production stress. Screens of gene perturbation libraries against production stressors can identify high-value engineering targets, but follow-up experiments needed to guard against false positives are slow and resource-intensive.
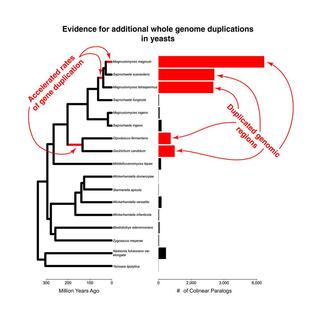
Discovery of additional ancient genome duplications in yeasts
Whole-genome duplication (WGD) has had profound macroevolutionary impacts on diverse lineages, preceding adaptive radiations in vertebrates, teleost fish, and angiosperms. In contrast to the many known ancient WGDs in animals, and especially plants, we are aware of evidence for only four WGDs in fungi.
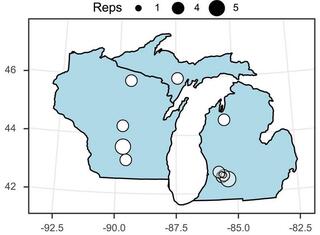
Switchgrass (Panicum virgatum) and miscanthus (Miscanthus × Giganteus) long-term yield patterns reveal consistent productivity declines
Perennial grasses like switchgrass (Panicum virgatum) and miscanthus (Miscanthus × giganteus) are expected to supply a substantial amount of the United States bioeconomy's feedstock demand. However, uncertainties around their long-term yields challenge the viability of their potential and limit their wider adoption.
Draft genome sequences of Burkholderia and Paraburkholderia strains isolated from Panicum virgatum soil and roots at the Lux Arbor Reserve in Michigan, USA
We present draft genomes of two Paraburkholderia strains and two Burkholderia strains. Three strains were isolated from surrounding soils and one from roots of switchgrass (Panicum virgatum) in Michigan, USA.
Stable hypermutators revealed by the genomic landscape of genes involved in genome stability among yeast species
Mutator phenotypes are short-lived due to the rapid accumulation of deleterious mutations. Yet, recent observations reveal that certain fungi can undergo prolonged accelerated evolution after losing genes involved in DNA repair.
Taxogenomic analysis of a novel yeast species, Lachancea rosae Sp. Nov. F.A., isolated from the wild rose Rosa californica
A novel Saccharomycotina yeast strain, yHQL494, was isolated from the rose hip of the wild rose Rosa californica from Castle Crags State Park, California, USA.
Draft genome of the switchgrass head smut pathogen Tilletia maclaganii
Tilletia maclaganii is a smut fungal pathogen that causes significant biomass reduction of switchgrass (Panicum virgatum) used for animal forage and biofuel production. Here we present the annotated genome of T. maclaganii, strain Tm001-NY21, estimated at 42.79 Mb in size, in 53 assembled contigs and encoding 10,235 predicted genes.
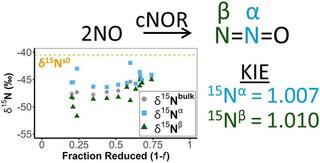
Isoptopic fractionation and kinetic isotope effects of a purified bacterial nitric oxide reductase (NOR)
Nitrous oxide (N2O) is a serious concern due to its role in global warming and ozone destruction. Agricultural practices account for ∼80% of all anthropogenic N2O produced in the US, due in large part to the stimulation of microbial denitrification.
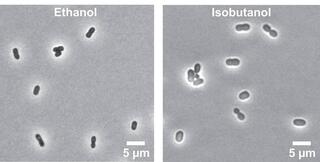
Physiological and metabolic responses of Zymomonas mobilis to lignocellulosic hydrolysate
Zymomonas mobilis is a promising biocatalyst for the sustainable conversion of lignocellulosic sugars into biofuels and bioproducts, yet its response to lignocellulosic hydrolysates remains poorly understood. Here, we investigate the physiological response of Z.
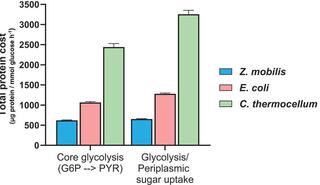
Thermodynamics shapes the in vivo enzyme burden on glycolytic pathways
Thermodynamically constrained reactions and pathways are hypothesized to impose greater protein demands on cells, requiring higher enzyme amounts to sustain a given flux compared to those with stronger thermodynamics.
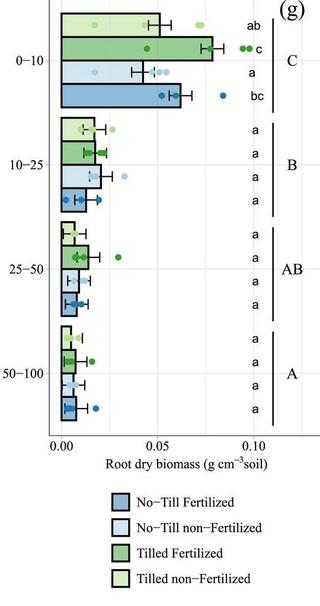
Agricultural management legacy effects on switchgrass growth and soil carbon gains
Switchgrass (Panicum virgatum L.) is a native North American grass currently considered a high-potential bioenergy feedstock crop. However, previous reports questioned its effectiveness in generating soil organic carbon (SOC) gains, with resultant uncertainty regarding the monoculture switchgrass's impact on the environmental sustainability of bioenergy agriculture.
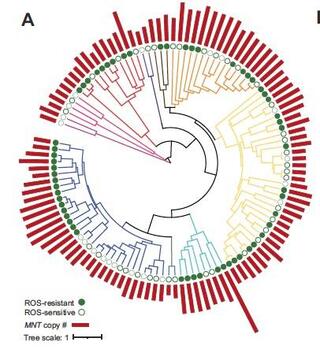
Machine learning reveals genes impacting oxidative stress resistance across yeasts
Reactive oxygen species (ROS) are highly reactive molecules encountered by yeasts during routine metabolism and during interactions with other organisms, including host infection.
MarK, a Novosphingobium aromaticivorans kinase required for catabolism of multiple aromatic monomers
The aromatic compounds used in a variety of industrial products are currently obtained from nonrenewable petroleum sources. Alternatively, the plant polymer lignin is an abundant renewable source of aromatics, and its depolymerization generates a variety of products that can include acetovanillone, a vanillin derivative containing an acetyl side chain.