An IRE1-proteasome system signalling cohort controls cell fate determination in unresolved proteotoxic stress of the plant endoplasmic reticulum
D.K. Ko et al. "An IRE1-proteasome system signalling cohort controls cell fate determination in unresolved proteotoxic stress of the plant endoplasmic reticulum" Nature Plants 9:1333 (2023) [DOI:10.1038/s41477-023-01480-3]
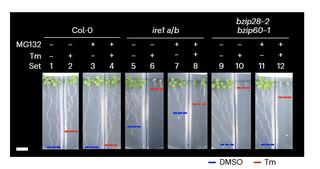
Excessive accumulation of misfolded proteins in the endoplasmic reticulum (ER) causes ER stress, which is an underlying cause of major crop losses and devastating human conditions. ER proteostasis surveillance is mediated by the conserved master regulator of the unfolded protein response (UPR), Inositol Requiring Enzyme 1 (IRE1), which determines cell fate by controlling pro-life and pro-death outcomes through as yet largely unknown mechanisms. Here we report that Arabidopsis IRE1 determines cell fate in ER stress by balancing the ubiquitin-proteasome system (UPS) and UPR through the plant-unique E3 ligase, PHOSPHATASE TYPE 2CA (PP2CA)-INTERACTING RING FINGER PROTEIN 1 (PIR1). Indeed, PIR1 loss leads to suppression of pro-death UPS and the lethal phenotype of an IRE1 loss-of-function mutant in unresolved ER stress in addition to activating pro-survival UPR. Specifically, in ER stress, PIR1 loss stabilizes ABI5, a basic leucine zipper (bZIP) transcription factor, that directly activates expression of the critical UPR regulator gene, bZIP60, triggering transcriptional cascades enhancing pro-survival UPR. Collectively, our results identify new cell fate effectors in plant ER stress by showing that IRE1's coordination of cell death and survival hinges on PIR1, a key pro-death component of the UPS, which controls ABI5, a pro-survival transcriptional activator of bZIP60.
All data supporting the findings of this study are available within this paper and its Supplementary Materials files. The raw data of WGS and RNA-seq have been deposited to the National Center for Biotechnological Information Sequence Read Archive and are accessible via BioProject accession codes PRJNA813856 and PRJNA880604. The Col-0 reference genome (TAIR10) was used for sequence analyses. The UPR eY1H dataset and DAP-seq dataset were downloaded from the corresponding database (https://brandizzilab.natsci.msu.edu/resources/upromics.aspx and http://neomorph.salk.edu/dev/pages/shhuang/dap_web/pages/index.php, respectively). The full results of WGS analyses are available in Supplementary Data 1. The processed data of RNA-seq analyses are available in Supplementary Data 2 and 3. Source data are provided with this paper. The scripts used in this study are available in GitHub (https://github.com/DaeKwan-Ko/ism93).