PKA regulatory subunit Bcy1 couples growth, lipid metabolism, and fermentation during anaerobic xylose growth in Saccharomyces cerevisiae
E.R. Wagner et al. "PKA regulatory subunit Bcy1 couples growth, lipid metabolism, and fermentation during anaerobic xylose growth in Saccharomyces cerevisiae" PLOS Genetics 19:e1010593 (2023) [DOI:10.1371/journal.pgen.1010593]
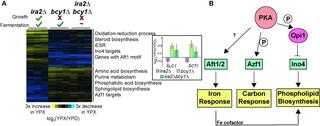
Organisms have evolved elaborate physiological pathways that regulate growth, proliferation, metabolism, and stress response. These pathways must be properly coordinated to elicit the appropriate response to an ever-changing environment. While individual pathways have been well studied in a variety of model systems, there remains much to uncover about how pathways are integrated to produce systemic changes in a cell, especially in dynamic conditions. We previously showed that deletion of Protein Kinase A (PKA) regulatory subunit BCY1 can decouple growth and metabolism in Saccharomyces cerevisiae engineered for anaerobic xylose fermentation, allowing for robust fermentation in the absence of division. This provides an opportunity to understand how PKA signaling normally coordinates these processes. Here, we integrated transcriptomic, lipidomic, and phospho-proteomic responses upon a glucose to xylose shift across a series of strains with different genetic mutations promoting either coupled or decoupled xylose-dependent growth and metabolism. Together, results suggested that defects in lipid homeostasis limit growth in the bcy1 delta strain despite robust metabolism. To further understand this mechanism, we performed adaptive laboratory evolutions to re-evolve coupled growth and metabolism in the bcy1 delta parental strain. The evolved strain harbored mutations in PKA subunit TPK1 and lipid regulator OPI1, among other genes, and evolved changes in lipid profiles and gene expression. Deletion of the evolved opi1 gene partially reverted the strain's phenotype to the bcy1 delta parent, with reduced growth and robust xylose fermentation. We suggest several models for how cells coordinate growth, metabolism, and other responses in budding yeast and how restructuring these processes enables anaerobic xylose utilization.
Raw and processed RNAseq files have been submitted to the NIH GEO database under project number GSE220465. Raw and processed lipidomic files were deposited in MassIVE database under dataset number MSV000090868