GLBRC Research Tools
On our research tools page, you can find a selection of web-based research tools and applications, developed collaboratively between GLBRC scientists and our Information Services (IS) group. If you are looking for a different tool or have any questions, contact IS Coordinator Dirk Norman (dirk.norman@wisc.edu).
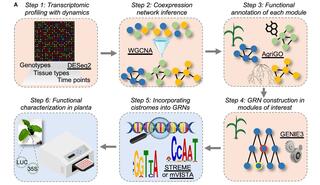
A network-enabled pipeline for gene discovery and validation in non-model plant species
Identifying key regulators of important genes in non-model crop species is challenging due to limited multi-omics resources.
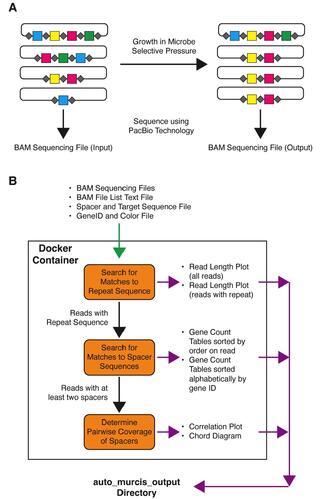
Auto-MuRCiS: a streamlined software package for analysis of multiplex, randomized CRISPR interference sequencing data
Multiplex, randomized CRISPR interference sequencing (MuRCiS) allows for the simultaneous identification of multiple gene knockouts that together influence microbial processes. Here, we report on an updated analysis tool called Auto-MuRCiS that utilizes Docker to make the analysis of these data rapid and more user-friendly.
Biomass Utilization Superstructure (BUS)
Biomass Utilization Superstructure (BUS) is an online tool for the evaluation of new and existing biofuels strategies. Users can modify and generate a new technology network that consists of a wide range of conversion technologies along with the corresponding feedstocks, intermediates, and final products.
COnTORT: COmprehensive Transcriptomic Organization Tool
COnTORT is a publicly available Python3 program that retrieves all available gene expression data and associated metadata for an organism from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database. The data are compiled into text files that can be used for downstream bioinformatic applications.
CRISpy-pop
CRISpy-pop is a web application that generates and filters guide RNA sequences for CRISPR/Cas9 genome editing. This tool focuses on generating guide RNA sequences for yeast and bacterial species used in bioenergy research. This tool supports a 1011 genome Saccharomyces cerevisiae strain set as well as a 167 strain set of S. cerevisiae isolates.
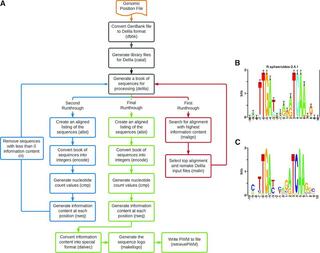
Delila-PY, a Pipeline for Utilizing the Delila Suite of Software to Identify Potential DNA Binding Motifs
Predicting potential DNA binding motifs is a critical part of understanding gene expression across all domains of life. Here, we report the development of Delila-PY, an easy-to-use pipeline for utilizing the Delila suite of software to identify DNA binding motifs. The Delila-PY Docker image can be found on the Docker hub.
Growth Curve Analysis Tool (GCAT)
The Growth Curve Analysis Tool is a web-based tool for summarizing microbial growth curves using mathematical modeling. The user interface requires no programming and calls on an R package of the same name which processes input data files, models the curves, calculates important growth parameters from the fits, and returns both graphical and tabular output.
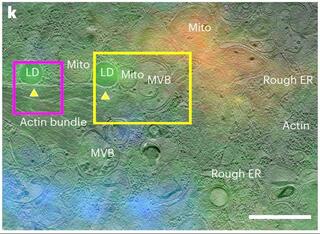
Integrated fluorescence light microscopy-guided cryo-focused ion beam-milling for in situ montage cryo-ET
Cryogenic-electron tomography (cryo-ET) permits the in situ visualization of biological macromolecules at the molecular level. Owing to the variable thickness of cells, tissues and organisms, frozen specimens may need to be thinned by cryo-focused ion beam (FIB) milling to produce thin (<500 nm) cryo-lamellae suitable for cryo-ET.
NMR Database of Lignin and Cell Wall Model Compounds
This is the latest public version of the Database, version 2024-08. Creating, maintaining, and updating this database is a huge job; please let us know if you find errors! We hope to have updates at least annually.
The database is in PDF format, providing the simplest way to get a viewable, printable, database on any platform, with limited searching capabilities.
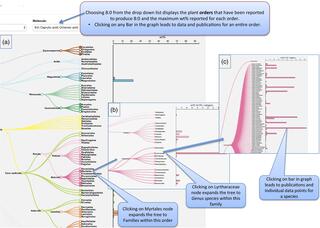
PlantFAdb: a resource for exploring hundreds of plant fatty acid structures synthesized by thousands of plants and their phylogenetic relationships
Over 450 structurally distinct fatty acids are synthesized by plants. We have developed PlantFAdb.org, an internet-based database that allows users to search and display fatty acid composition data for over 9000 plants. PlantFAdb includes more than 17 000 data tables from >3000 publications and hundreds of unpublished analyses.
sppIDer
Yeast 1000 Plus (Y1000+)
The Y1000+ project is funded by the National Science Foundation to determine "The Making of Biodiversity Across the Yeast Subphylum." Over the next five years, the team behind the project will sequence and analyze the genomes of all known yeast species from the subphylum Saccharomycotina to study the evolution of their diverse metabolic and ecological functions.