Uncovering a miltiradiene biosynthetic gene cluster in the Lamiaceae reveals a dynamic evolutionary trajectory
A.E. Bryson et al. "Uncovering a miltiradiene biosynthetic gene cluster in the Lamiaceae reveals a dynamic evolutionary trajectory" Nature Communications 14:343 (2023) [DOI:10.1038/s41467-023-35845-1]
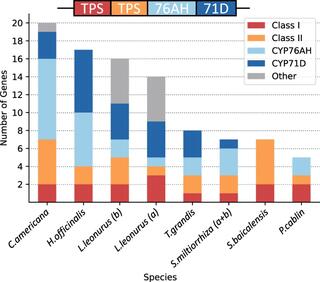
The spatial organization of genes within plant genomes can drive evolution of specialized metabolic pathways. Terpenoids are important specialized metabolites in plants with diverse adaptive functions that enable environmental interactions. Here, we report the genome assemblies of Prunella vulgaris, Plectranthus barbatus, and Leonotis leonurus. We investigate the origin and subsequent evolution of a diterpenoid biosynthetic gene cluster (BGC) together with other seven species within the Lamiaceae (mint) family. Based on core genes found in the BGCs of all species examined across the Lamiaceae, we predict a simplified version of this cluster evolved in an early Lamiaceae ancestor. The current composition of the extant BGCs highlights the dynamic nature of its evolution. We elucidate the terpene backbones generated by the Callicarpa americana BGC enzymes, including miltiradiene and the terpene (+)-kaurene, and show oxidization activities of BGC cytochrome P450s. Our work reveals the fluid nature of BGC assembly and the importance of genome structure in contributing to the origin of metabolites.
The data supporting the findings of this work are available within the paper and the Supplementary Information files. The raw genomic reads generated in this study have been deposited in the NCBI BioSample database under the following accession codes: Plectranthus barbatus (SAMN26547115), Leonotis leonurus (SAMN26547116), and Prunella vulgaris (SAMN26547117). The genome assemblies have been deposited in NCBI with the following accession codes: Plectranthus barbatus (JAPKLW000000000 [https://www.ncbi.nlm.nih.gov/nuccore/JAPKLW000000000.1/]), Leonotis leonurus (JAPKLX000000000 [https://www.ncbi.nlm.nih.gov/nuccore/JAPKLX000000000.1/]), and Prunella vulgaris (JAPKLY000000000 [https://www.ncbi.nlm.nih.gov/nuccore/JAPKLY000000000.1/]). The versions described in this paper are versions XXXXXX010000000. Sequences for the functionally characterized enzymes from Callicarpa americana can be found in the NCBI GenBank database: ON260868 [https://www.ncbi.nlm.nih.gov/nuccore/ON260868.1/]-ON260876 [https://www.ncbi.nlm.nih.gov/nuccore/ON260876.1/]. Additional data including genome assemblies and annotations, GC-MS raw data, NMR raw data, phylogenetic alignments, cluster sequences, and collinearity files can be found in our Dryad Repository. Source data are provided with this paper.