Lignin deconstruction by anaerobic fungi
T.S. Lankiewicz et al. "Lignin deconstruction by anaerobic fungi" Nature Microbiology 8:596-610 (2023) [DOI:10.1038/s41564-023-01336-8]
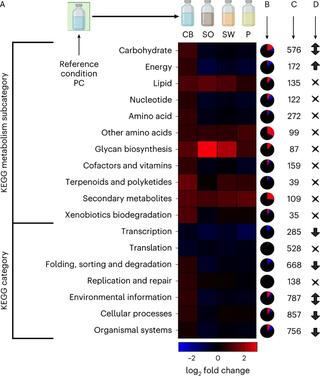
Lignocellulose forms plant cell walls, and its three constituent polymers, cellulose, hemicellulose and lignin, represent the largest renewable organic carbon pool in the terrestrial biosphere. Insights into biological lignocellulose deconstruction inform understandings of global carbon sequestration dynamics and provide inspiration for biotechnologies seeking to address the current climate crisis by producing renewable chemicals from plant biomass. Organisms in diverse environments disassemble lignocellulose, and carbohydrate degradation processes are well defined, but biological lignin deconstruction is described only in aerobic systems. It is currently unclear whether anaerobic lignin deconstruction is impossible because of biochemical constraints or, alternatively, has not yet been measured. We applied whole cell-wall nuclear magnetic resonance, gel-permeation chromatography and transcriptome sequencing to interrogate the apparent paradox that anaerobic fungi (Neocallimastigomycetes), well-documented lignocellulose degradation specialists, are unable to modify lignin. We find that Neocallimastigomycetes anaerobically break chemical bonds in grass and hardwood lignins, and we further associate upregulated gene products with the observed lignocellulose deconstruction. These findings alter perceptions of lignin deconstruction by anaerobes and provide opportunities to advance decarbonization biotechnologies that depend on depolymerizing lignocellulose.
All data are available in the main text, the Extended Data, the Supplementary Information or through JGl’s Mycocosm (https://mycocosm.jgi.doe.gov/mycocosm/ home). Raw reads of differential expression data are deposited in NCBl’s Sequence Read Archive database and can be found with SRA accession numbers SRP288871–SRP288885. Amplicon sequences for fungal taxonomic classification are deposited in GenBank under BioProject accession number PRJNA800048.