Codon optimization improves the prediction of xylose metabolism from gene content in budding yeasts
R.L. Nalabothu et al. "Codon optimization improves the prediction of xylose metabolism from gene content in budding yeasts" Molecular Biology and Evolution 40:msad111 (2023) [DOI:10.1093/molbev/msad111]
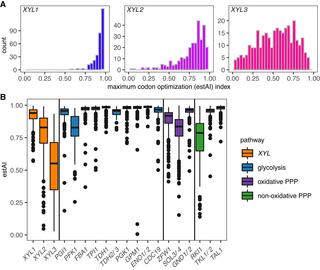
Xylose is the second most abundant monomeric sugar in plant biomass. Consequently, xylose catabolism is an ecologically important trait for saprotrophic organisms, as well as a fundamentally important trait for industries that hope to convert plant mass to renewable fuels and other bioproducts using microbial metabolism. Although common across fungi, xylose catabolism is rare within Saccharomycotina, the subphylum that contains most industrially relevant fermentative yeast species. The genomes of several yeasts unable to consume xylose have been previously reported to contain the full set of genes in the XYL pathway, suggesting the absence of a gene-trait correlation for xylose metabolism. Here, we measured growth on xylose and systematically identified XYL pathway orthologs across the genomes of 332 budding yeast species. Although the XYL pathway coevolved with xylose metabolism, we found that pathway presence only predicted xylose catabolism about half of the time, demonstrating that a complete XYL pathway is necessary, but not sufficient, for xylose catabolism. We also found that XYL1 copy number was positively correlated, after phylogenetic correction, with xylose utilization. We then quantified codon usage bias of XYL genes and found that XYL3 codon optimization was significantly higher, after phylogenetic correction, in species able to consume xylose. Finally, we showed that codon optimization of XYL2 was positively correlated, after phylogenetic correction, with growth rates in xylose medium. We conclude that gene content alone is a weak predictor of xylose metabolism and that using codon optimization enhances the prediction of xylose metabolism from yeast genome sequence data.
Analyses were performed on the 332 published and publicly available assemblies analyzed in Shen et al. (2018). Codon optimization values were obtained from the figshare repository from LaBella et al. (2019) (https://doi.org/10.6084/m9.figshare.c.4498292). All data generated in this project, including curated XYL gene sequences, are available in the figshare associated with this manuscript (https://doi.org/10.6084/m9.figshare.c.6011956.v1).