GLBRC Data Sets
Highlighted below are a variety of published studies that include data sets that might be of interest to the scientific community and have been deposited in online data repositories. Only data sets published in GLBRC-approved repositories following the FAIR Guiding Principles are highlighted. More information can be found on our guidelines page.
Mixed acid fermentation of carbohydrate-rich dairy manure hydrolysate
A.T. Ingle et al. "Mixed acid fermentation of carbohydrate-rich dairy manure hydrolysate" Frontiers in Bioengineering and Biotechnology 9 (2021) [DOI: 10.3389/fbioe.2021.724304]
The aim of this study was to investigate if carbon derived from DM fibers can be recovered as medium-chain fatty acids (MCFAs), which are mixed culture fermentation products of economic interest. DM fibers were subjected to combinations of physical, enzymatic, chemical, and thermochemical pretreatments to evaluate the possibility of producing carbohydrate-rich hydrolysates suitable for microbial fermentation by mixed cultures.
Diverse profile of fermentation byproducts from thin stillage
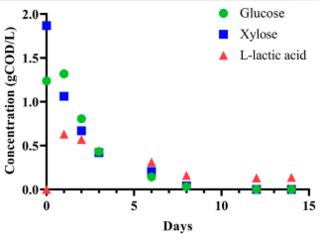
N.W. Fortney et al. "Diverse profile of fermentation byproducts from thin stillage" Frontiers in Bioengineering and Biotechnology (2021) 9 [DOI: 10.3389/fbioe.2021.695306]
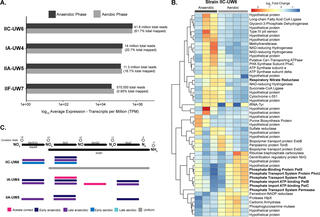
Here, we explored the formation of different fermentation products by microbial communities grown on TS using different bioreactor conditions. At the baseline operational condition (6-day retention time, pH 5.5, 35°C), we observed a mixture of MCFAs as the principal fermentation products.
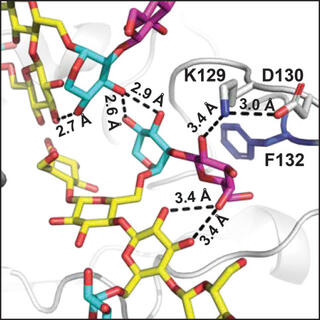
A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis
E.M. Glasgow et al. "A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis" Journal of Biological Chemistry 295, 51 (2020) [10.1074/jbc.RA120.015328]
A mechanistic understanding of GH5_4 substrate specificity would help inform the best protein design strategies and the most appropriate industrial application of broad-specificity endoglucanases. Here we report structures of 10 new GH5_4 enzymes from cellulolytic microbes and characterize their substrate selectivity using normalized reducing sugar assays and MS.
Geographic variation in the genetic basis of resistance to leaf rust between locally adapted ecotypes of the biofuel crop switchgrass (Panicum virgatum)
A. VanWallendael et al. "Geographic variation in the genetic basis of resistance to leaf rust between locally adapted ecotypes of the biofuel crop switchgrass (Panicum virgatum)" New Phytologist 227 (2020) [DOI: 10.1111/nph.16555]
To understand how the genetic architecture of resistance varies across time and geographic space, we quantified rust (Puccinia spp.) severity in switchgrass (Panicum virgatum) plantings at eight locations across the central USA for 3 yr and conducted quantitative trait locus (QTL) mapping for rust progression.
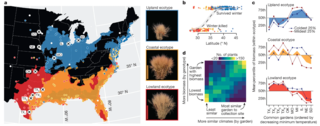
Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass
J.T. Lovell et al. "Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass" Nature 590 (2021) [DOI: 10.1038/s41586-020-03127-1]
Here we present the assembly and annotation of the large and complex genome of the polyploid bioenergy crop switchgrass (Panicum virgatum). Analysis of biomass and survival among 732 resequenced genotypes, which were grown across 10 common gardens that span 1,800 km of latitude, jointly revealed extensive genomic evidence of climate adaptation.
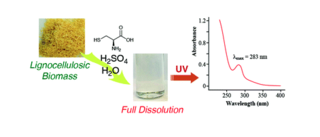
A facile spectroscopic method for measuring lignin content in lignocellulosic biomass
F. Lu et al. "A facile spectroscopic method for measuring lignin content in lignocellulosic biomass" Green Chemistry 14 (2021) [DOI: 10.1039/D1GC01507A]
A facile spectroscopic method, our CASA (Cysteine–Assisted Sulfuric Acid) method, was developed to quantify the lignin content of lignocellulosic biomass, based on an extraordinary system in which biomass samples are fully dissolved in H2SO4.
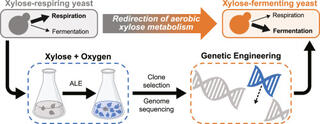
Crabtree/Warburg-like aerobic xylose fermentation by engineered Saccharomyces cerevisiae
S. Lee et al. "Crabtree/Warburg-like aerobic xylose fermentation by engineered Saccharomyces cerevisiae" Metabolic Engineering 68 (2021) [DOI: 10.1016/j.ymben.2021.09.008]
Here, we evolved respiration-deficient S. cerevisiae strains that can grow on and ferment xylose to ethanol aerobically, a trait analogous to the Crabtree/Warburg Effect for glucose.
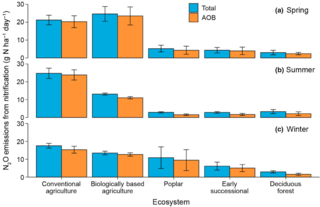
Nitrification is a minor source of nitrous oxide in an agricultural landscape and declines with increasing management intensity
D. Liang and G.P. Robertson "Nitrification is a minor source of nitrous oxide (N2O) in an agricultural landscape and declines with increasing management intensity" Global Change Biology 27, 21 (2021) [DOI: 10.1111/gcb.15833]
Using Bayesian inference to couple 25 years of in situ N2O flux measurements with site-specific Michaelis–Menten kinetics of nitrification-derived N2O, we test the relative importance of nitrification-derived N2O across six cropped and unmanaged ecosystems along a management intensity gradient in the U.S. Midwest.
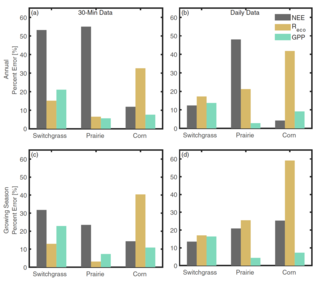
Modeled surface-atmosphere fluxes from paired sites in the Upper Great Lakes Region using neural networks
D.E. Reed et al. "Modeled surface-atmosphere fluxes from paired sites in the Upper Great Lakes Region using neural networks" JGR Biogeosciences 126, 8 (2021) [DOI: 10.1029/2021JG006363]
Here we use a long-term record of carbon cycling from six paired sites plus a seventh reference site that are all spatially close to each other, with an average of ∼11 km separation. The goal is to use field observations from one of the paired sites, along with observations from the second site that can be measured easily, in order to estimate carbon cycling at the second site using artificial intelligence methods.
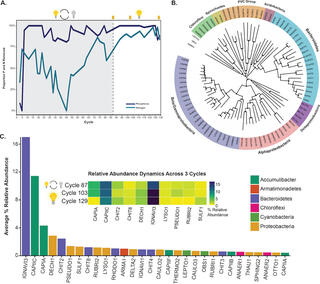
Metabolic differentiation of co-occurring Accumulibacter clades revealed through genome-resolved metatranscriptomics
E.A. McDaniel et al. "Metabolic differentiation of co-occurring Accumulibacter clades revealed through genome-resolved metatranscriptomics" Environmental Microbiology 6, 4 (2021) [DOI: 10.1128/mSystems.00474-21]
Here, we performed time series genome-resolved metatranscriptomics to explore gene expression patterns of coexisting Accumulibacter clades in the same bioreactor ecosystem. Our work provides an approach for elucidating ecologically relevant functions based on gene expression patterns between closely related microbial populations.
Hanseniaspora smithiae sp. nov., a novel apiculate yeast species from Patagonian forests that lacks the typical genomic domestication signatures for fermentative environments
N. Čadež et al. "Hanseniaspora smithiae sp. nov., a novel apiculate yeast species from Patagonian forests that lacks the typical genomic domestication signatures for fermentative environments" Frontiers in Microbiology (2021) [DOI: 10.3389/fmicb.2021.679894]
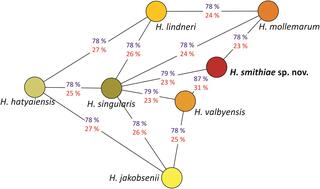
During a survey of Nothofagus trees and their parasitic fungi in Andean Patagonia (Argentina), genetically distinct strains of Hanseniaspora were obtained from the sugar-containing stromata of parasitic Cyttaria spp.
Contributions of environmental and maternal transmission to the assembly of leaf fungal endophyte communities
L.P. Bell-Dereske and S.E. Evans "Contributions of environmental and maternal transmission to the assembly of leaf fungal endophyte communities" Proceedings of the Royal Society B 288, 1956 (2021) [DOI: 10.1098/rspb.2021.0621]
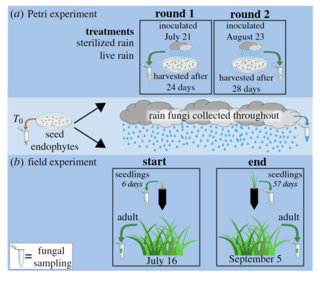
We used amplicon sequencing to track switchgrass (Panicum virgatum) LFEs in a greenhouse and field experiment as communities assembled from seed endophytes and rain fungi (integration of wet and dry aerial dispersal) in germinating seeds, seedlings, and adult plants.
Natural variation in the consequences of gene overexpression and its implications for evolutionary trajectories
D. Robinson et al. "Natural variation in the consequences of gene overexpression and its implications for evolutionary trajectories" eLife (2021) [DOI: 10.7554/eLife.70564]
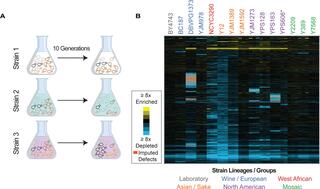
Here, we measured fitness costs of over 4000 overexpressed genes in 15 Saccharomyces cerevisiae strains representing different lineages, to explore natural variation in tolerating gene overexpression (OE).
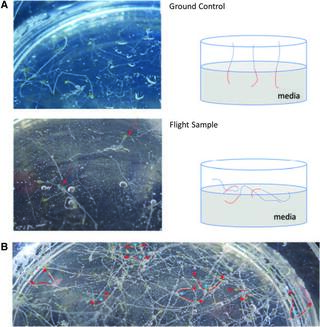
Relevance of the unfolded protein response to spaceflight-induced transcriptional reprogramming in Arabidopsis
E. Angelos et al. “Relevance of the unfolded protein response to spaceflight-induced transcriptional reprogramming in Arabidopsis” Astrobiology 21, 3 (2021) [DOI: 10.1089/ast.2020.2313]
To contribute to the understanding of how plants respond to spaceflight stress, we examined the significance of the unfolded protein response (UPR), a conserved signaling cascade that responds to a number of unfavorable environmental stresses, in the model plant Arabidopsis thaliana.
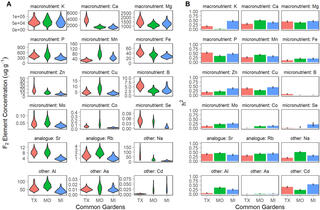
QTL x environment interactions underlie ionome divergence in switchgrass
L. Zhang et al. "QTL x environment interactions underlie ionome divergence in switchgrass" G3 Genes|Genomes|Genetics (2021) [DOI:10.1093/g3journal/jkab144]
We grew 725 clonally replicated genotypes of a large full sib family from a four-way linkage mapping population, created from deeply diverged upland and lowland switchgrass ecotypes, at three common gardens.
A multi-omics approach to lignocellulolytic enzyme discovery reveals a new ligninase activity from Parascedosporium putredinis NO1
N.C. Oates et al. "A multi-omics approach to lignocellulolytic enzyme discovery reveals a new ligninase activity from Parascedosporium putredinis NO1" PNAS 118,18 (2021) [DOI:10.1073/pnas.2008888118]
Here, we describe the isolation of an exceptional lignocellulose-degrading fungus that produces new oxidase activity with no cofactor requirements.
Genome-resolved metagenomics of a photosynthetic bioreactor performing biological nutrient removal
E.A. McDaniel et al. "Genome-resolved metagenomics of a photosynthetic bioreactor performing biological nutrient removal" Microbiology Resource Announcements 10,18 (2021) [DOI:10.1128/MRA.00244-21]
We present a genome-resolved metagenomics data set consisting of 86 metagenome-assembled genome sequences from a photosynthetically operated lab-scale bioreactor simulating EBPR.
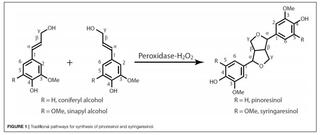
Efficient synthesis of pinoresinol, an important lignin dimeric model compound
F. Yue et al. "Efficient synthesis of pinoresinol, an important lignin dimeric model compound" Frontiers in Energy Research 9 (2021) [DOI: 10.3389/fenrg.2021.640337]
The synthetic yield and isolation efficiency of racemic pinoresinol from coniferyl alcohol by conventional radical coupling methods is sub-optimal. In this work, a facile and efficient synthetic approach was developed to synthesize pinoresinol with much higher yield.
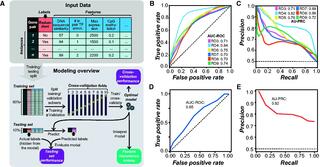
Predictive models of genetic redundancy in Arabidopsis thaliana
S.A. Cusack et al. "Predictive models of genetic redundancy in Arabidopsis thaliana" Molecular Biology and Evolution (2021) [DOI: 10.1093/molbev/msab111]
Here, we establish machine learning models for predicting whether a gene pair is likely redundant or not in the model plant Arabidopsis thaliana based on six feature categories: functional annotations, evolutionary conservation including duplication patterns and mechanisms, epigenetic marks, protein properties including posttranslational modifications, gene expression, and gene network properties.
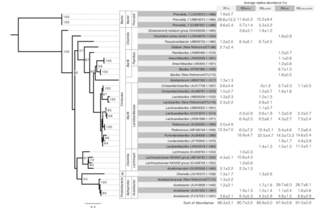
A genome-scale phylogeny of the kingdom Fungi
L. Yuanning et al. “A genome-scale phylogeny of the kingdom Fungi” Nature Ecology 31, 8 (2020) [DOI:10.1016/j.cub.2021.01.074]
To examine poorly resolved relationships and assess progress toward a genome-scale phylogeny of the fungal kingdom, we compiled a phylogenomic data matrix of 290 genes from the genomes of 1,644 species that includes representatives from most major fungal lineages.