Hanseniaspora smithiae sp. nov., a novel apiculate yeast species from Patagonian forests that lacks the typical genomic domestication signatures for fermentative environments
N. Čadež et al. "Hanseniaspora smithiae sp. nov., a novel apiculate yeast species from Patagonian forests that lacks the typical genomic domestication signatures for fermentative environments" Frontiers in Microbiology (2021) [DOI: 10.3389/fmicb.2021.679894]
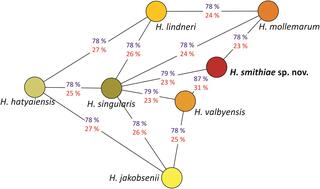
During a survey of Nothofagus trees and their parasitic fungi in Andean Patagonia (Argentina), genetically distinct strains of Hanseniaspora were obtained from the sugar-containing stromata of parasitic Cyttaria spp. Phylogenetic analyses based on the single-gene sequences (encoding rRNA and actin) or on conserved, single-copy, orthologous genes from genome sequence assemblies revealed that these strains represent a new species closely related to Hanseniaspora valbyensis. Additionally, delimitation of this novel species was supported by genetic distance calculations using overall genome relatedness indices (OGRI) between the novel taxon and its closest relatives. To better understand the mode of speciation in Hanseniaspora, we examined genes that were retained or lost in the novel species in comparison to its closest relatives. These analyses show that, during diversification, this novel species and its closest relatives, H. valbyensis and Hanseniaspora jakobsenii, lost mitochondrial and other genes involved in the generation of precursor metabolites and energy, which could explain their slower growth and higher ethanol yields under aerobic conditions. Similarly, Hanseniaspora mollemarum lost the ability to sporulate, along with genes that are involved in meiosis and mating. Based on these findings, a formal description of the novel yeast species Hanseniaspora smithiae sp. nov. is proposed, with CRUB 1602H as the holotype.
The datasets presented in this study can be found in MycoBank under number MB839330. The GenBank accession numbers of datasets used for the analysis can be found in Supplementary Table S1.