Metabolic differentiation of co-occurring Accumulibacter clades revealed through genome-resolved metatranscriptomics
E.A. McDaniel et al. "Metabolic differentiation of co-occurring Accumulibacter clades revealed through genome-resolved metatranscriptomics" Environmental Microbiology 6, 4 (2021) [DOI: 10.1128/mSystems.00474-21]
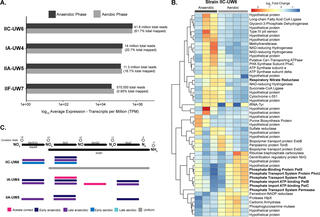
“Candidatus Accumulibacter phosphatis” is a model polyphosphate-accumulating organism that has been studied using genome-resolved metagenomics, metatranscriptomics, and metaproteomics to understand the EBPR process. Within the Accumulibacter lineage, several similar but diverging clades are defined by the shared sequence identity of the polyphosphate kinase (ppk1) locus. These clades are predicted to have key functional differences in acetate uptake rates, phage defense mechanisms, and nitrogen-cycling capabilities. However, such hypotheses have largely been made based on gene content comparisons of sequenced Accumulibacter genomes, some of which were obtained from different systems. Here, we performed time series genome-resolved metatranscriptomics to explore gene expression patterns of coexisting Accumulibacter clades in the same bioreactor ecosystem. Our work provides an approach for elucidating ecologically relevant functions based on gene expression patterns between closely related microbial populations.
Raw sequencing files for the three metagenomes and seven metatranscriptomes and genome assemblies for the four Accumulibacter genomes are available in the NCBI database under BioProject accession number PRJNA668760. All genome assemblies used in this study in their current forms as well as transcriptomic count tables are available on figshare. All code is available on GitHub.