QTL x environment interactions underlie ionome divergence in switchgrass
L. Zhang et al. "QTL x environment interactions underlie ionome divergence in switchgrass" G3 Genes|Genomes|Genetics (2021) [DOI:10.1093/g3journal/jkab144]
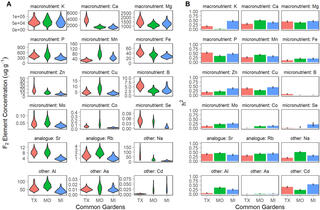
Ionomics measures elemental concentrations in biological organisms and provides a snapshot of physiology under different conditions. In this study, we evaluate genetic variation of the ionome in outbred, perennial switchgrass in three environments across the species’ native range, and explore patterns of genotype-by-environment interactions. We grew 725 clonally replicated genotypes of a large full sib family from a four-way linkage mapping population, created from deeply diverged upland and lowland switchgrass ecotypes, at three common gardens. Concentrations of 18 mineral elements were determined in whole post-anthesis tillers using ion coupled plasma mass spectrometry (ICP-MS). These measurements were used to identify quantitative trait loci (QTL) with and without QTL-by-environment interactions (QTLxE) using a multi-environment QTL mapping approach. We found that element concentrations varied significantly both within and between switchgrass ecotypes, and GxE was present at both the trait and QTL level. Concentrations of 14 of the 18 elements were under some genetic control, and 77 QTL were detected for these elements. 74% of QTL colocalized multiple elements, half of QTL exhibited significant QTLxE, and roughly equal numbers of QTL had significant differences in magnitude and sign of their effects across environments. The switchgrass ionome is under moderate genetic control and by loci with highly variable effects across environments.
The supplemental materials are available at figshare.