GLBRC Data Sets
Highlighted below are a variety of published studies that include data sets that might be of interest to the scientific community and have been deposited in online data repositories. Only data sets published in GLBRC-approved repositories following the FAIR Guiding Principles are highlighted. More information can be found on our guidelines page.
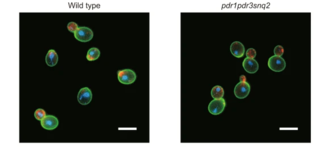
High-throughput platform for yeast morphological profiling predicts the targets of bioactive compounds
S. Ohnuki et al. "High-throughput platform for yeast morphological profiling predicts the targets of bioactive compounds" npj Systems Biology and Applications 8 (2022) [DOI: 10.1038/s41540-022-00212-1]
In this study, we developed a reliable high-throughput (HT) platform for yeast morphological profiling using drug-hypersensitive strains to minimize compound use, HT microscopy to speed up data generation and analysis, and a generalized linear model to predict targets with high reliability.
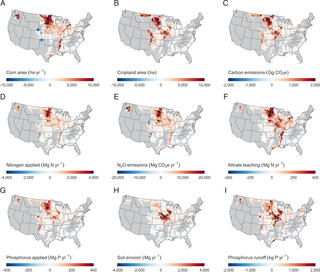
Environmental outcomes of the US Renewable Fuel Standard
Lark et al. "Environmental outcomes of the US Renewable Fuel Standard" PNAS 119 (2022) [DOI: 10.1073/pnas.2101084119]
Biofuels are included in many proposed strategies to reduce anthropogenic greenhouse gas emissions and limit the magnitude of global warming. The US Renewable Fuel Standard is the world’s largest existing biofuel program, yet despite its prominence, there has been limited empirical assessment of the program’s environmental outcomes.
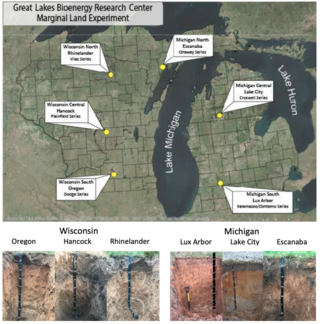
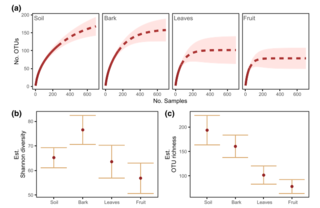
Soils of the GLBRC Marginal Land Expeirment (MLE) Sites
C.S. Kasmerchak and R. Schaetzl "Soils of the GLBRC Marginal Land Experiment (MLE) Sites" KBS LTER Special Publication (2018) [DOI: 10.5281/zenodo.2578238]
The Great Lakes Bioenergy Research Center (GLBRC) Marginal Land Experiment (MLE) was established in 2013 at various sites in Wisconsin and Michigan to evaluate the potential use of low productivity or abandoned agricultural fields for low input bioenergy feedstocks.
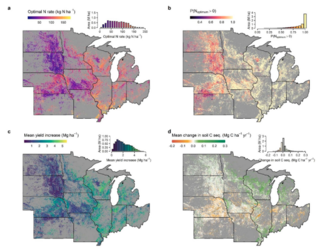
Boosting climate change mitigation potential of perennial lignocellulosic crops grown on marginal lands
R.A. Martinez-Feria and B. Basso "Boosting climate change mitigation potential of perennial lignocellulosic crops grown on marginal lands" Environmental Research Letters 17 (2022) [DOI: 10.1088/1748-9326/ac541b]
Here, we show the potential GHG mitigation of fertilizing switchgrass (Panicum virgatum) at the NF rate that minimizes net GHG emissions across 7.1 million ha of marginal lands in the Midwest US, with long-term production advantages surpassing emitted GHG by 0.66 Mg CO2e ha−1 yr−1 on the aggregate.
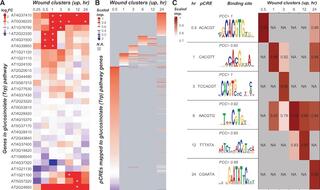
Modeling temporal and hormonal regulation of plant transcriptional response to wounding
Moore et al. "Modeling temporal and hormonal regulation of plant transcriptional response to wounding" The Plant Cell (2021) [DOI:10.1093/plcell/koab287]
Using a combination of genome editing, in vitro DNA-binding assays, and transient expression assays using native and mutated cis-regulatory elements, we experimentally validated four of the predicted elements, three of which were not previously known to function in wound-response regulation.
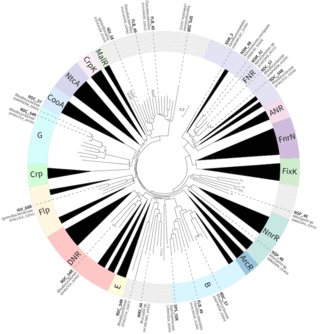
Exploring the meta-regulon of the CRP/FNR family of global transcriptional regulators in a partial-nitritation anammox microbiome
N.K. Beach et al. "Exploring the meta-regulon of the CRP/FNR family of global transcriptional regulators in a partial-nitritation anammox microbiome" mSystems 6 (2021) [DOI:10.1128/mSystems.00906-21]
Optimizing cyclic aeration helps reduce energy needs and maximize microbiome performance during wastewater treatment; however, little is known about how most microbial community members respond to these alternating conditions. We defined the meta-regulon of a PNA microbiome by combining existing knowledge of how the CRP/FNR family of bacterial TFs respond to stimuli, with metatranscriptomic analyses to characterize gene expression changes during aeration cycles.
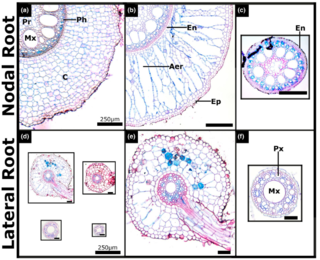
Bioenergy sorghum’s deep roots: A key to sustainable biomass production on annual cropland
A. Lamb et al. "Bioenergy sorghum’s deep roots: A key to sustainable biomass production on annual cropland" GCB-Bioenergy 14 (2021) [DOI:10.1111/gcbb.12907]
In this study, field grown bioenergy sorghum root systems were analyzed during the growing season to characterize their depth, biomass, morphology, anatomy, and gene expression profiles. Bioenergy sorghum roots grew continuously during a 155-day growing season producing ~175 nodal roots, accumulating ~7 Mg of dry biomass per hectare, and reaching >2 m deep in the soil profile.
Investigating the chemolithoautotrophic and formate metabolism of Nitrospira moscoviensis by constraint-based metabolic modeling and 13C-tracer analysis
C.E. Lawson et al. "Investigating the Chemolithoautotrophic and Formate Metabolism of Nitrospira moscoviensis by Constraint-Based Metabolic Modeling and 13C-Tracer Analysis" mSystems 6 (2021) [DOI:10.1128/mSystems.00173-21]
We provide the first constraint-based metabolic model of Nitrospira moscoviensis representing the ubiquitous Nitrospira lineage II and subsequently validate this model using proteomics and 13C-tracers combined with intracellular metabolomic analysis. The resulting genome-scale model will serve as a knowledge base of Nitrospira metabolism and lays the foundation for quantitative systems biology studies of these globally important nitrite-oxidizing bacteria.
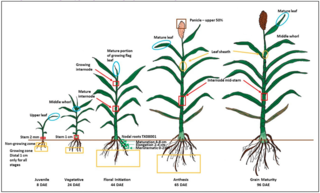
Regulation of dhurrin pathway gene expression during Sorghum bicolor development
R.M. Gleadow et al. "Regulation of dhurrin pathway gene expression during Sorghum bicolor development" Planta 254 (2021) [DOI:10.1007/s00425-021-03774-2]
In this study, RNA-seq was used to investigate the expression of genes involved in the biosynthesis, bio-activation and recycling of dhurrin in Sorghum bicolor. Genes involved in dhurrin biosynthesis were highly expressed in all young developing vegetative tissues (leaves, leaf sheath, roots, stems), tiller buds and imbibing seeds and showed gene specific peaks of expression in leaves during diel cycles.
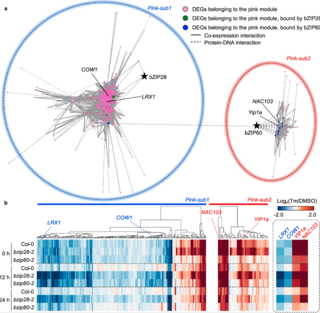
Advanced genomics identifies growth effectors for proteotoxic ER stress recovery in Arabidopsis thaliana
D.K. Ko and F. Brandizzi "Advanced genomics identifies growth effectors for proteotoxic ER stress recovery in Arabidopsis thaliana" Communications Biology 5 (2022) [DOI:10.1038/s42003-021-02964-8]
We undertook a genomics approach in the model plant species Arabidopsis thaliana and mined the gene reprogramming roles of the UPR modulators, basic leucine zipper28 (bZIP28) and bZIP60, in ER stress resolution. Through a network modeling and experimental validation, we identified key genes downstream of the UPR bZIP-transcription factors (bZIP-TFs), and demonstrated their functional roles.
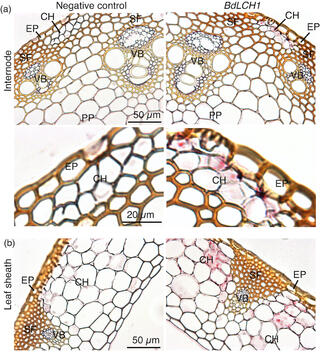
Disruption of Brachypodium lichenase alters metabolism of mixed-linkage glucan and starch
M. Fan et al. "Disruption of Brachypodiumlichenase alters metabolism of mixed-linkage glucan and starch" The Plant Journal (2021) [DOI:10.1111/tpj.1560312]
We identify a gene encoding a lichenase we name Brachypodium distachyon LICHENASE 1 (BdLCH1), which is highly expressed in the endosperm of germinating seeds and coleoptiles and at lower amounts in mature shoots. RNA in situ hybridization showed that BdLCH1 is primarily expressed in chlorenchyma cells of mature leaves and internodes.
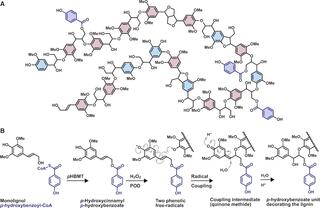
pHBMT1, a BAHD-family monolignol acyltransferase, mediates lignin acylation in poplar
de Vries et al. "pHBMT1, a BAHD-family monolignol acyltransferase, mediates lignin acylation in poplar" Plant Physiology (2021) [DOI:10.1093/plphys/kiab546]
We performed an in vitro screen of the Populus trichocarpa BAHD acyltransferase superfamily (116 genes) using a wheatgerm cell-free translation system and found five enzymes capable of producing monolignol–p-hydroxybenzoates. We then compared the transcript abundance of the five corresponding genes with p-hydroxybenzoate concentrations using naturally occurring unrelated genotypes of P. trichocarpa.
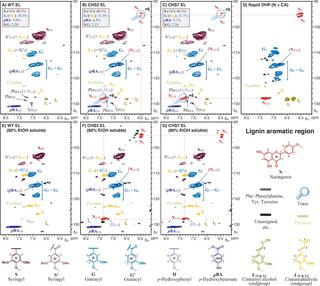
Exogenous chalcone synthase expression in developing poplar xylem incorporates naringenin into lignins
E.L. Mahon et al. "Exogenous chalcone synthase expression in developing poplar xylem incorporates naringenin into lignins" Plant Physiology 188 (2021) [DOI:doi.org/10.1093/plphys/kiab499]
We engineered hybrid poplar (Populus alba x grandidentata) to express chalcone synthase 3 (MdCHS3) derived from apple (Malus domestica) in lignifying xylem. Transgenic trees displayed an accumulation of the flavonoid naringenin in xylem methanolic extracts not inherently observed in wild-type trees.
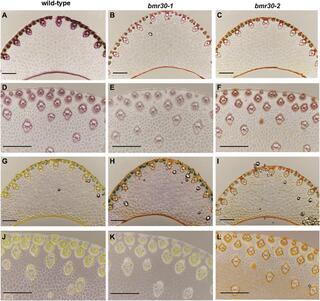
The sorghum (Sorghum bicolor) brown midrib 30 gene encodes a chalcone isomerase required for cell wall lignification
H.M. Tetreault et al. "The Sorghum (Sorghum bicolor) Brown Midrib 30 Gene Encodes a Chalcone Isomerase Required for Cell Wall Lignification" Frontiers in Plant Science (2021) [DOI:10.3389/fpls.2021.732307]
In sorghum (Sorghum bicolor) and other C4 grasses, brown midrib (bmr) mutants have long been associated with plants impaired in their ability to synthesize lignin. The brown midrib 30 (Bmr30) gene, identified using a bulk segregant analysis and next-generation sequencing, was determined to encode a chalcone isomerase (CHI).
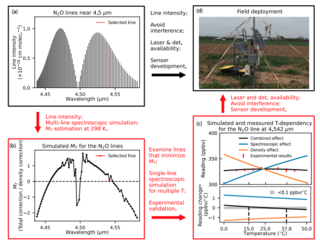
A new open-path eddy covariance method for nitrous oxide and other trace gases that minimizes temperature corrections
D. Pan et al. "A new open-path eddy covariance method for nitrous oxide and other trace gases that minimizes temperature corrections" Global Change Biology 28 (2021) [DOI:10.1111/gcb.15986]
We demonstrate a new laser-based, open-path N2O sensor and a general approach applicable to other gases that minimizes temperature-related corrections for EC flux measurements. The method identifies absorption lines with spectroscopic effects in the opposite direction of density effects from temperature and, thus, density and spectroscopic effects nearly cancel one another.
Substrate, temperature, and geographical patterns among nearly 2,000 natural yeast isolates
W.J. Spurley et al. "Substrate, temperature, and geographical patterns among nearly 2,000 natural yeast isolates" Yeast (2021) [DOI:10.1002/yea.3679]
We used environmental sampling and isolation to assemble a dataset of 1962 isolates collected from throughout the contiguous United States of America (USA) and Alaska, which were then used to uncover geographic patterns, along with substrate and temperature associations among yeast taxa.
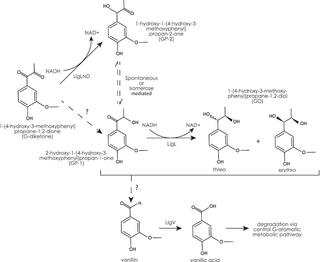
Aromatic dimer dehydrogenases from Novosphingobium aromaticivorans reduce monoaromatic diketones
A.M. Linz et al. "Aromatic dimer dehydrogenases from Novosphingobium aromaticivorans reduce monoaromatic diketones" Applied and Enviromental Microbiology 87 (2021) [DOI:10.1128/AEM.01742-21]
In this work, we report a newly discovered activity of previously characterized dehydrogenase enzymes with a chemically modified by-product of lignin depolymerization. We propose that the activity of N. aromaticivorans enzymes with both native lignin aromatics and those produced by chemical depolymerization will expand opportunities for producing industrial chemicals from the heterogenous components of this abundant plant polymer.
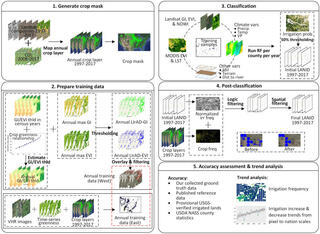
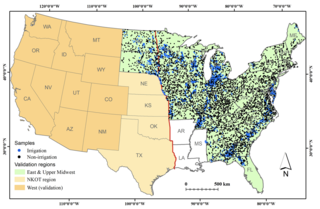
Mapping annual irrigation from Landsat imagery and environmental variables across the conterminous United States
Y. Xie et al. "Mapping annual irrigation from Landsat imagery and environmental variables across the conterminous United States" Remote Sensing of Environment 260 (2021) [DOI: 10.1016/j.rse.2021.112445]
In this study, we present an approach to map the extent of irrigated croplands across the conterminous U.S. (CONUS) for each year in the period of 1997–2017. To scale nationwide, we developed novel methods to generate training datasets covering both the western and eastern U.S.
Landsat-based Irrigation Dataset (LANID): 30 m resolution maps of irrigation distribution, frequency, and change for the US, 1997–2017
Y. Xie et al. "Landsat-based Irrigation Dataset (LANID): 30-m resolution maps of irrigation distribution, frequency, and change for the U.S., 1997–2017" Earth System Science Data 13 (2021) [DOI:10.5194/essd-13-5689-2021]
Here we present the new Landsat-based Irrigation Dataset (LANID), which is comprised of 30 m resolution annual irrigation maps covering the conterminous US (CONUS) for the period of 1997–2017. The main dataset identifies the annual extent of irrigated croplands, pastureland, and hay for each year in the study period. Derivative maps include layers on maximum irrigated extent, irrigation frequency and trends, and identification of formerly irrigated areas and intermittently irrigated lands.
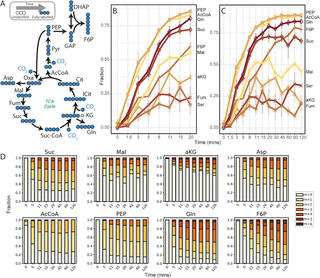
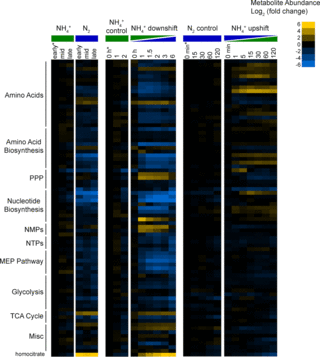
Metabolic remodeling during nitrogen fixation in Zymomonas mobilis
J.I. Martien et al. "Metabolic remodeling during nitrogen fixation in Zymomonas mobilis" mSystems 6, 6 (2021) [DOI: 10.1128/mSystems.00987-21]
In Z. mobilis, the factors that control glycolytic rates, ethanol production, and isoprenoid production are still not fully understood. In this study, we performed metabolomic, proteomic, and thermodynamic analysis of Z. mobilis during N2 fixation. This analysis identified key changes in metabolite levels, enzyme abundance, and glycolytic thermodynamic favorability that occurred during changes in NH4+ availability, helping to inform future efforts in metabolic engineering.