Investigating the chemolithoautotrophic and formate metabolism of Nitrospira moscoviensis by constraint-based metabolic modeling and 13C-tracer analysis
C.E. Lawson et al. "Investigating the Chemolithoautotrophic and Formate Metabolism of Nitrospira moscoviensis by Constraint-Based Metabolic Modeling and 13C-Tracer Analysis" mSystems 6 (2021) [DOI:10.1128/mSystems.00173-21]
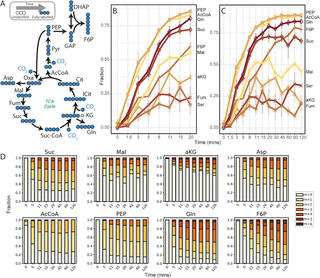
Nitrospira spp. are globally abundant nitrifying bacteria in soil and aquatic ecosystems and in wastewater treatment plants, where they control the oxidation of nitrite to nitrate. Despite their critical contribution to nitrogen cycling across diverse environments, detailed understanding of their metabolic network and prediction of their function under different environmental conditions remains a major challenge. Here, we provide the first constraint-based metabolic model of Nitrospira moscoviensis representing the ubiquitous Nitrospira lineage II and subsequently validate this model using proteomics and 13C-tracers combined with intracellular metabolomic analysis. The resulting genome-scale model will serve as a knowledge base of Nitrospira metabolism and lays the foundation for quantitative systems biology studies of these globally important nitrite-oxidizing bacteria.
The transcriptomic data set reanalyzed here can be found under the NCBI Gene Expression Omnibus. The mass spectrometry proteomics raw data is available via the PRIDE repository.