Exploring the meta-regulon of the CRP/FNR family of global transcriptional regulators in a partial-nitritation anammox microbiome
N.K. Beach et al. "Exploring the meta-regulon of the CRP/FNR family of global transcriptional regulators in a partial-nitritation anammox microbiome" mSystems 6 (2021) [DOI:10.1128/mSystems.00906-21]
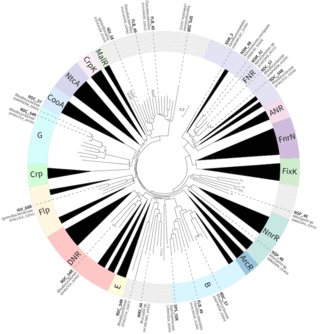
Microbiomes are important contributors to many ecosystems, including ones where nutrient cycling is stimulated by aeration control. Optimizing cyclic aeration helps reduce energy needs and maximize microbiome performance during wastewater treatment; however, little is known about how most microbial community members respond to these alternating conditions. We defined the meta-regulon of a PNA microbiome by combining existing knowledge of how the CRP/FNR family of bacterial TFs respond to stimuli, with metatranscriptomic analyses to characterize gene expression changes during aeration cycles. Our results indicated that, for some members of the community, prior knowledge is sufficient for high-confidence assignments of TF function, whereas other community members have CRP/FNR TFs for which inferences of function are limited by lack of prior knowledge. This study provides a framework to begin elucidating meta-regulons in microbiomes, where pure cultures are not available for traditional transcriptional regulation studies. Defining the meta-regulon can help in optimizing microbiome performance.
MAGs and annotations are available on the NCBI BioProject. DNA sequencing reads are available via the NCBI SRA. RNA sequencing reads are available through the NCBI SRA.