GLBRC Data Sets
Highlighted below are a variety of published studies that include data sets that might be of interest to the scientific community and have been deposited in online data repositories. Only data sets published in GLBRC-approved repositories following the FAIR Guiding Principles are highlighted. More information can be found on our guidelines page.
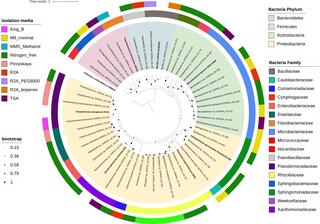
Genome-sequenced bacterial collection from sorghum aerial root mucilage
A collection of 47 bacteria isolated from the mucilage of aerial roots of energy sorghum is available at the Great Lakes Bioenergy Research Center, Michigan State University, Michigan, USA. We enriched bacteria with putative plant-beneficial phenotypes and included information on phenotypic diversity, taxonomy, and whole genome sequences.
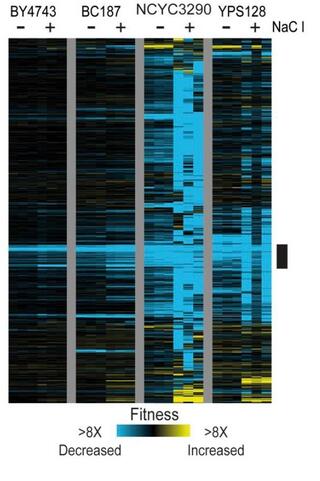
Gene-by-environment interactions influence the fitness cost of gene copy-number variation in yeast
Variation in gene copy number can alter gene expression and influence downstream phenotypes; thus copy-number variation (CNV) provides a route for rapid evolution if the benefits outweigh the cost.
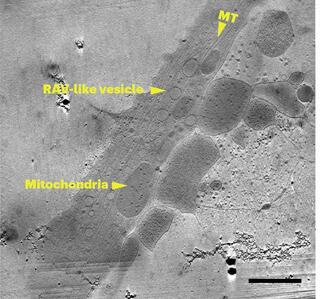
Correlative montage parallel array cryo-tomography for in situ structural cell biology
Imaging large fields of view while preserving high-resolution structural information remains a challenge in low-dose cryo-electron tomography. Here we present robust tools for montage parallel array cryo-tomography (MPACT) tailored for vitrified specimens.
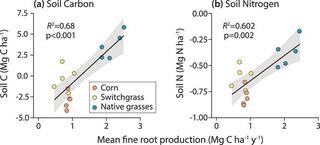
Long-term changes in soil carbon and nitrogen fractions in switchgrass, native grasses, and no-till corn bioenergy production system
Cellulosic bioenergy is a primary land-based climate mitigation strategy, with soil carbon (C) storage and nitrogen (N) conservation as important mitigation elements. Here, we present 13 years of soil C and N change under three cellulosic cropping systems: monoculture switchgrass (Panicum virgatum L.), a five native grasses polyculture, and no-till corn (Zea mays L.).
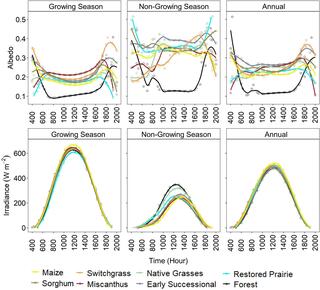
Climate cooling benefits of cellulosic bioenergy crops from elevated albedo
Changes in land surface albedo can alter ecosystem energy balance and potentially influence climate.
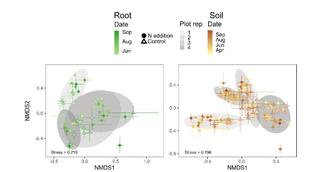
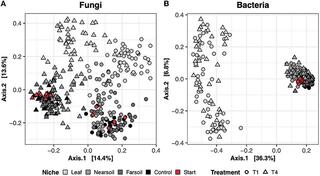
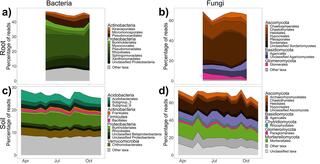
Regional biogeography versus intra-annual dynamics of the root and soil microbiome
Root and soil microbial communities constitute the below-ground plant microbiome, are drivers of nutrient cycling, and affect plant productivity. However, our understanding of their spatiotemporal patterns is confounded by exogenous factors that covary spatially, such as changes in host plant species, climate, and edaphic factors.
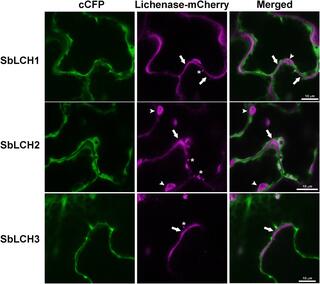
Cell- and development-specific degradation controls the levels of mixed-linkage glucan in sorghum leaves
Mixed-linkage glucan (MLG) is a component of the cell wall (CW) of grasses and is composed of glucose monomers linked by beta-1,3 and beta-1,4 bonds. MLG is believed to have several biological functions, such as the mobilizable storage of carbohydrates and structural support of the CW.
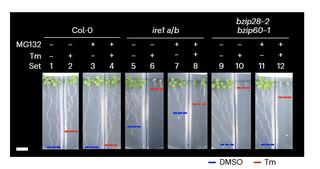
An IRE1-proteasome system signalling cohort controls cell fate determination in unresolved proteotoxic stress of the plant endoplasmic reticulum
Excessive accumulation of misfolded proteins in the endoplasmic reticulum (ER) causes ER stress, which is an underlying cause of major crop losses and devastating human conditions.
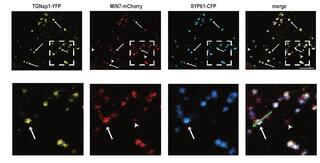
Defense against phytopathogens relies on efficient antimicrobial protein secretion mediated by the microtubule-binding protein TGNap1
Plant immunity depends on the secretion of antimicrobial proteins, which occurs through yet-largely unknown mechanisms. The trans-Golgi network (TGN), a hub for intracellular and extracellular trafficking pathways, and the cytoskeleton, which is required for antimicrobial protein secretion, are emerging as pathogen targets to dampen plant immunity.
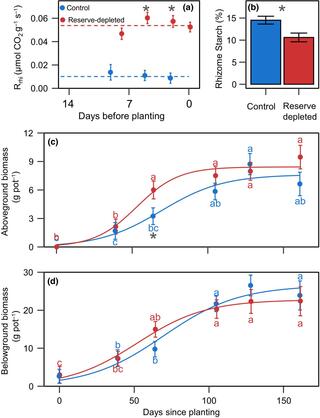
A 30% reduction in switchgrass rhizome reserves did not decrease biomass yield
A long-standing question in perennial grass breeding and physiology is whether yield improvement strategies could compromise winter survival. Since perennial grasses rely on stored carbohydrates for winter maintenance and spring regrowth, yield improvement strategies could reduce winter survival if they increase biomass and grain yields at the expense of carbon allocation to storage.
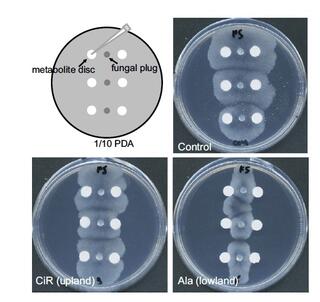
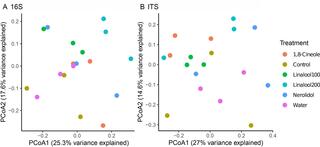
Anti-fungal bioactive terpenoids in the bioenergy crop switchgrass (Panicum virgatum) may contribute to ecotype-specific microbiome composition
Plant derived bioactive small molecules have attracted attention of scientists across fundamental and applied scientific disciplines. We seek to understand the influence of these phytochemicals on rhizosphere and root-associated fungi.
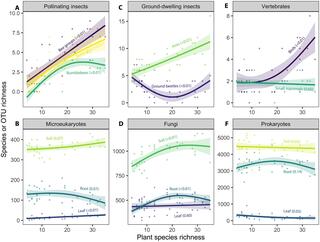
Contrasting effects of bioenergy crops on biodiversity
Agriculture is driving biodiversity loss, and future bioenergy cropping systems have the potential to ameliorate or exacerbate these effects. Using a long-term experimental array of 10 bioenergy cropping systems, we quantified diversity of plants, invertebrates, vertebrates, and microbes in each crop.
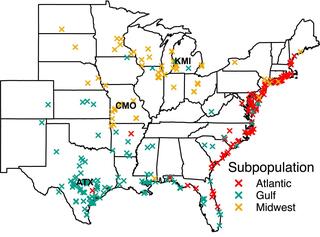
Genetic determinants of switchgrass-root-associated microbiota in field sites spanning its natural range
A fundamental goal in plant microbiome research is to determine the relative impacts of host and environmental effects on root microbiota composition, particularly how host genotype impacts bacterial community composition.
Terpenes modulate bacterial and fungal growth and sorghum rhizobiome communities
Terpenes represent one of the largest and oldest classes of plant-specialized metabolism, but their role in the belowground microbiome is poorly understood.
The microbiome structure of decomposing plant leaves in soil depends on plant species, soil pore sizes, and soil moisture content
Microbial communities are known as the primary decomposers of all the carbon accumulated in the soil. However, how important soil structure and its conventional or organic management, moisture content, and how different plant species impact this process are less understood.
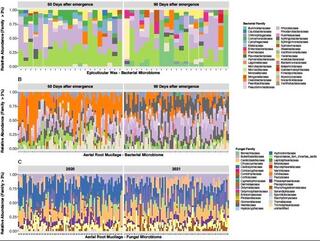
Phyllosphere exudates select for distinct microbiome members in sorghum epicuticular wax and aerial root mucilage
Phyllosphere exudates create specialized microhabitats that shape microbial community diversity. We explored the microbiome associated with two sorghum phyllosphere exudates, the epicuticular wax and aerial root mucilage.
Regional biogeography versus intra-annual dynamics of the root and soil microbiome
Root and soil microbial communities constitute the below-ground plant microbiome, are drivers of nutrient cycling, and affect plant productivity. However, our understanding of their spatiotemporal patterns is confounded by exogenous factors that covary spatially, such as changes in host plant species, climate, and edaphic factors.
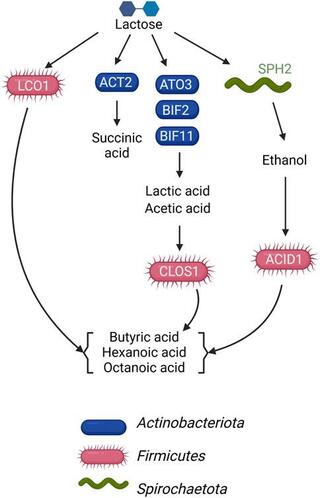
A metagenome-level analysis of a microbial community fermenting ultra-filtered milk permeate
Fermentative microbial communities have the potential to serve as biocatalysts for the conversion of low-value dairy coproducts into renewable chemicals, contributing to a more sustainable global economy.
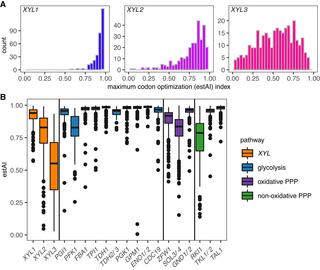
Codon optimization improves the prediction of xylose metabolism from gene content in budding yeasts
Xylose is the second most abundant monomeric sugar in plant biomass. Consequently, xylose catabolism is an ecologically important trait for saprotrophic organisms, as well as a fundamentally important trait for industries that hope to convert plant mass to renewable fuels and other bioproducts using microbial metabolism.
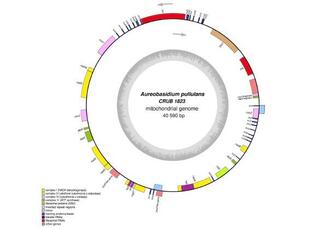
Assembly and comparative genome analysis of a Patagonian Aureobasidium pullulans isolate reveals unexpected intraspecific variation
Aureobasidium pullulans is a yeast-like fungus with remarkable phenotypic plasticity widely studied for its importance for the pharmaceutical and food industries. So far, genomic studies with strains from all over the world suggest they constitute a genetically unstructured population, with no association by habitat.