Genome-sequenced bacterial collection from sorghum aerial root mucilage
M.E. Mechan-Llontop et al. "Genome-sequenced bacterial collection from sorghum aerial root mucilage" Microbiology Resource Announcements 12:e00468-23 (2023) [DOI:10.1128/MRA.00468-23]
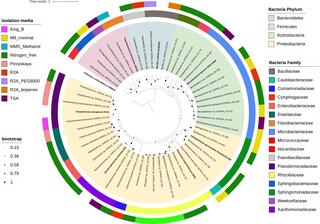
A collection of 47 bacteria isolated from the mucilage of aerial roots of energy sorghum is available at the Great Lakes Bioenergy Research Center, Michigan State University, Michigan, USA. We enriched bacteria with putative plant-beneficial phenotypes and included information on phenotypic diversity, taxonomy, and whole genome sequences.
Raw sequencing data are available at the Joint Genome Institute’s GOLD database under Study ID Gs0157305, as well as with the National Institutes of Health Sequence Read Archive (see Table 1). Genome sequencing assembly data have been deposited in the GenBank with accession numbers listed in Table 1. The bacterial collection is available at The Great Lakes Bioenergy Research Center at Michigan State University, USA (glbrc@msu.edu).