Research Highlights

Great Lakes Bioenergy researchers and collaborators engineered softwoods to incorporate a key feature of hardwoods. The resulting pine (shown here) processes more easily into pulp and paper.
Great Lakes Bioenergy research consistently results in new discoveries and new technologies. Here, we highlight high-impact research from all three of our research areas.
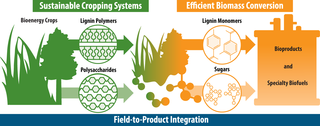
Outlining an integrated pipeline from biomass to bioproduct
A new study from GLBRC describes a complete lignin-to-bioproduct pipeline that could produce high yields of a target chemical, 2-pyrone-4,6-dicarboxylic acid (PDC), with high potential value as a platform chemical to make adhesives, plastics, and other biopolymers.
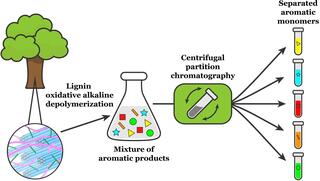
Isolating valuable compounds from complex mixtures of lignin products
In a new study, a team from the Great Lakes Bioenergy Research Center describes a multi-solvent extraction process that can isolate five major products obtained from poplar lignin.
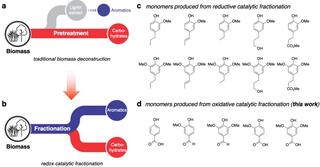
Simplifying biomass breakdown toward making biofuels and chemicals
Chemists at the Great Lakes Bioenergy Research Center (GLBRC) have developed a new approach that converts biomass into both carbohydrates and lignin-derived aromatic monomers in a single step. This approach avoids lignin degradation and mass losses that occur in traditional biomass separation methods and enables better product yields.
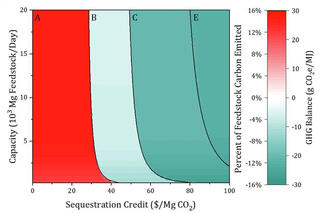
Optimizing for carbon storage: a new model for biorefineries
GLBRC scientists incorporated carbon capture and storage technologies into models of how crop and pretreatment choices influence the economic viability of a biofuel refinery. They developed a model and performed thousands of individual calculations to determine the impact of carbon capture and storage technologies, feedstock choices, and pretreatment strategies on three outcomes: energy production, environmental impacts, economic benefits.
Bioenergy crops create local cooling that can increase climate benefit
A research team at the Great Lakes Bioenergy Research Center has developed a way to measure GWI associated with albedo, which calculates how well a crop’s surface reflectivity cools the local climate. A new study shows that albedo-induced climate mitigation from conversion of CRP lands to perennial bioenergy crops like switchgrass and restored prairie can be substantial.
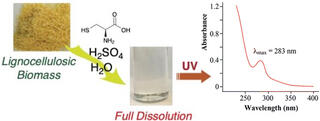
A safer, more efficient way to measure lignin content in biomass
Researchers at GLBRC and the South China University of Technology have developed a faster and less chemically intensive spectroscopy-based procedure called the Cysteine-Assisted Sulfuric Acid (CASA) method. The approach uses a new reagent combination of the amino acid cysteine and sulfuric acid to completely dissolve lignocellulosic biomass under less extreme conditions than traditional methods.
When it comes to soil carbon storage, a plant’s neighbors can make all the difference
A key discovery by Great Lakes Bioenergy Research Center scientists reveals how root interactions among Midwest prairie perennials, including the bioenergy crop switchgrass, increase soil carbon when grown in specific pairings. These beneficial plant combinations also created more soil pores that support fungi that are associated with soil carbon accrual.
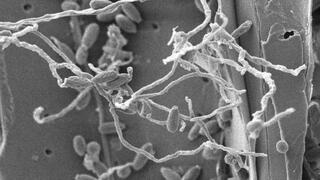
Fungus yields a new biomass-digesting enzyme
A team from the University of York and the Great Lakes Bioenergy Research Center isolated a soft-rot fungus that shows exceptional ability to break down lignocellulose. From this fungus, they identified oxidase enzyme activity that effectively cleaves one of the major bonds in lignin, a process that currently requires expensive and time-consuming pretreatment steps. The enzyme also releases potentially useful co-products and makes the remaining lignin more easily digested by other enzymes.
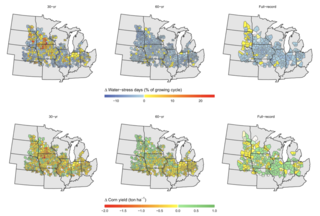
Planning for crop water needs under changing climate conditions
As the climate warms, many scientists and farmers have worried about how rising temperatures will affect agricultural systems and crop yields. Warmer air may increase evaporation and water loss from plants, leading to concerns that climate change will cause crops to require more water—either from precipitation or irrigation—in a warmer future. However, a modeling study using temperature trends projected through 2050 suggests that future yields of corn and other bioenergy crops in the Midwest may stay stable without need for expanded irrigation.
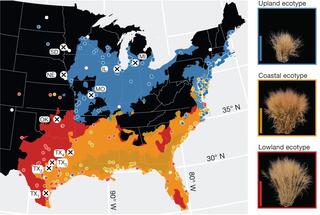
Nationwide collaboration unlocks switchgrass genome
In the face of climate change, breeding strategies that can improve crop resilience and productivity across a range of environments is increasingly important, but these approaches require sufficient knowledge of the genes that underlie productivity and adaptation. By studying 732 genotypes grown at 10 experimental gardens in eight states spread across 1,100 miles, a large team of researchers have produced a high-quality reference sequence of the complex switchgrass genome.