Machine learning models guide genetics research
Interpreting the logic behind machine learning models trained to analyze complex datasets can answer research questions in genetics and genomics.
The Science
Machine learning (ML) is a powerful tool for finding and analyzing relationships in large, complex biological datasets. ML works by identifying patterns in data and using those patterns to create models that can make predictions about new data. Although powerful, ML models cannot be easily understood by humans due to their complexity. Luckily, strategies to demystify the logic of ML models are available and continually improving. These model interpretation strategies are already being applied to genetics and genomics datasets to extract information about underlying biological processes. That information can then be used to generate specific hypotheses for experimental testing.
The Impact
The ability to understand the logic driving an ML model is key for learning from the model and trusting its predictions. Advances in interpretable ML are allowing researchers to identify features and relationships in large genetics and genomics datasets that may be challenging or even impossible to analyze with other approaches. As ML approaches continue to increase in computational complexity, it is becoming even more important for computational biologists to contribute novel interpretation strategies. The challenges and potential of interpretable ML also highlight the importance of training the next generation of biologists to work at the intersection of computer and biological sciences.
Summary
ML uses computer algorithms to find patterns in complex data, making it a critical tool for scientists to make sense of nonlinear, multi-dimensional biological datasets, such as multi-omics. For example, scientists at the Great Lakes Bioenergy Research Center are developing ML models to determine how to enhance switchgrass productivity by genetically modifying flowering time traits. Such models would speed up the breeding process by quickly identifying varieties with the desirable flowering time traits based solely on their genes.
ML models are developed using a dataset composed of instances (e.g., individual switchgrass plants) and features describing those instances (e.g., genetic variants of switchgrass). Given a label, or variable of interest (such as flowering time) for some of the instances, an ML model can be trained to predict unknown labels.
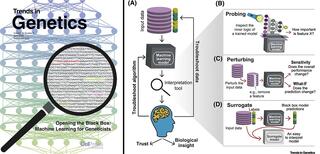
Model interpretation helps explore associations between the input features and the output predictions. The three major ML interpretation strategies are probing, perturbing, and surrogate strategies. Probing inspects a model’s parameters to see what the model has learned and how important each feature is in the model’s predictions. Perturbing involves changing values of an input feature and measuring the change in the model’s performance to understand that feature’s contribution. For models that cannot be probed or perturbed, a surrogate strategy can be used to train a second, inherently more interpretable, ML model to approximate the predictions of the first one. As described in the paper, many user-friendly tools are available to facilitate interpreting ML models using these strategies.
Associations identified through ML interpretation can then be tested experimentally. In bioenergy research, for example, ML models will be used to speed up the introduction of desirable traits to extend feedstock growing seasons, analyze below-ground microbiome diversity, and predict fitness of microbial strains with different genetic backgrounds in a wide range of biorefinery hydrolysates.
Program Manager
N. Kent Peters
kent.peters@science.doe.gov, 301-903-5549
Corresponding Author
Shin-Han Shiu
shius@msu.edu, (517) 353-7196
Funding
Funding was provided in part by the Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under award numbers DE-SC0018409, and by National Science Foundation Graduate Research Fellowship (Fellow ID: 2015196719), and NSF grants numbers IIS-1907704, IIS-1845081, CNS-1815636, IOS-1546617, DEB-1655386.
Publications
C.B. Azodi, et al., “Opening the Black Box: Interpretable Machine Learning for Geneticists.” Trends in Genetics 36, 442-455 (2020). [DOI: https://doi.org/10.1016/j.tig.2020.03.005]
Related Links
https://www.cell.com/trends/genetics/fulltext/S0168-9525(20)30069-X
https://www.glbrc.org/news/how-geneticists-can-use-machine-learning