NEEDLE points to key gene regulators
Background
Meeting the energy and food demands of the growing world population requires the rapid development of climate change-resilient crops, which scientists can produce by reprogramming genes that control growth, development, and stress responses. Multi-omic datasets are often limited to model plant species, but as information becomes increasingly available, complex datasets could be processed with new computational networks.
Approach
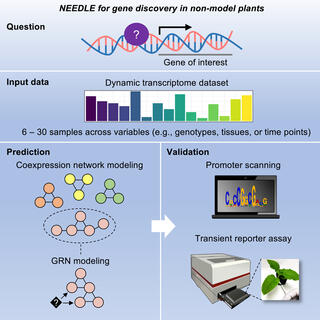
Scientists with the Great lakes Bioenergy Research Center designed NEEDLE, a user-friendly platform to delineate regulatory interactions within plants, and tested it on both known and unknown questions in order to determine its accuracy and usefulness in identifying transcription factors of target genes.
Results
NEEDLE identified transcription factors regulating the expression of the crucial cell wall biosynthetic gene Cellulose Synthase-Like F6 (CSLF6) in Brachypodium and sorghum and validated their regulatory function. The analyses not only uncovered novel regulators of CSLF6 but revealed both functional divergence and conservation of TFs across species through cross-species prediction and validation or divergence of gene regulatory elements among grass species.
Impact
This network requires a minimal input of data compared to other bioinformatic pipelines and is designed with modeling procedures that can be adapted and improved. Overall, NEEDLE represents an efficient framework to aid the bioengineering of crops, particularly in non-model plant species with available dynamic transcriptome datasets.