Natural genetic diversity of yeast aids in identification of genes involved in ionic liquid (IL) tolerance
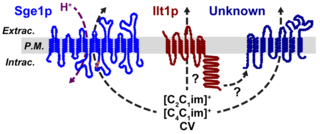
Mining yeast genomes for tolerance to ILs yields insight into cellular defense mechanisms and strategies to improve strain performance.
The Science
Microbes from nature represent a range of biological variation that can be exploited to identify traits that contribute to a particular phenotype. In this study, researchers examined natural diverse isolates of Saccaromyces cerevisiae for tolerance to ILs and identified two genes associated with this outcome. Analysis of specific gene variants suggests that tolerance may be governed in part by the degree to which ILs can be pumped out of cells.
The Impact
An understanding of the cellular mechanisms of IL tolerance can enable rational engineering approaches to design robust microbial strains that efficiently convert biomass to biofuels. The range of natural biological variation present among diverse microbial isolates is an important resource that can contribute to identification of biological mechanisms of interest.
Summary
ILs are promising deconstruction solvents for the conversion of lignocellulosic biomass to biofuels. However, many conversion microbes are sensitive to the toxic effects of residual solvent. One approach to circumventing this problem is to identify genetic traits that contribute to IL tolerance and engineer them into biofuel-producing strains for improved performance. Researchers examined the growth of 136 S. cerevisiae genome-sequenced strains in media containing 1-ethyl-3-methyl imidazolium chloride ([C2C1im]Cl) in order to gauge natural phenotypic variation among yeast from diverse ecological niches. The best performing strain was analyzed and compared to an IL-sensitive strain to determine the genetic basis for tolerance. From a screened library of genomic DNA fragments, two genes were associated with improved IL tolerance: SGE1, which encodes a plasma membrane multidrug efflux pump, and a previously uncharacterized gene (designated here as ILT1), encoding a predicted membrane protein. Comparison of SGE1 sequences across strains implicated two single nucleotide polymorphisms (SNPs) that associated with IL tolerance and sensitivity. The phenotypic effects of the SNPs were confirmed by CRISPR/Cas9 genome editing of a [C2C1im]Cl-sensitive strain. Interestingly, the SNPs were determined to affect Sge1 protein stability and cell surface localization, potentially impacting the amount of toxic ILs that cells can pump out of the cytoplasm. These results demonstrate the wealth of biological function inherent in nature that may be exploited for biocatalyst strain improvement and provide further clues on the cellular mechanisms of IL tolerance.
Contacts (BER PM)
N. Kent Peters
Program Manager, Office of Biological and Environmental Research
kent.peters@science.doe.gov, 301-903-5549
(PI Contact)
Trey K. Sato
Great Lakes Bioenergy Research Center
University of Wisconsin–Madison
tksato@glbrc.wisc.edu
Funding
This material is partly based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0018409, and work funded by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494). This work was also part of the DOE Joint BioEnergy Institute, supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy.
Publications
Higgins, D.A. et al., “Natural variation in the multidrug efflux pump SGE1 underlies ionic liquid tolerance in yeast.” Genetics (2018), DOI: 10.1534/genetics.118.301161.