Discovery of a new type of bacterial enzyme able to cleave bonds in lignin
Diversity in b-etherase pathways could help to valorize lignin
The Science
Characterizing microbial strategies for lignin breakdown is important for understanding plant biomass turnover in nature, and could aid in developing industrial systems for producing commodity chemicals from this abundant renewable resource. In this report, Great Lakes Bioenergy Research Center researchers provide new information on the pathway used by sphingomonad bacteria to cleave the β-aryl ether bond commonly found in lignin. Specifically, they report on a previously uncharacterized heterodimeric β-etherase enzyme, with unique properties, from Novosphingobium aromaticivorans and other sphingomonads.
The Impact
Lignocellulosic plant biomass is one of the most plentiful organic materials on Earth, so deciphering the mechanisms of its decomposition and harnessing this activity may provide strategies for recovering valuable bioproducts from lignin. Great Lakes Bioenergy Research Center researchers have identified and characterized a new type of enzyme that specifically cleaves β-aryl ether bonds, the most prevalent type of linkage found in the lignin polymer. This work demonstrates that there is more variability in the bacterial pathway for cleaving the β-aryl ether bond than previously thought, and illustrates the importance of studying this pathway – as well as other pathways involved in metabolizing lignin-derived compounds – in multiple species to better understand and be able to capitalize on this diversity.
Summary
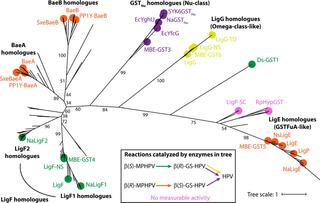
Lignin is a heterogeneous polymer of aromatic subunits that is a major component of lignocellulosic plant biomass. Understanding how microorganisms deconstruct lignin is important for understanding the global carbon cycle, and could aid in developing systems for processing plant biomass into valuable commodities. Sphingomonad bacteria use stereospecific glutathione S-transferases (GSTs) called β-etherases to cleave the β-aryl ether (β-O-4) bond, the most common bond between aromatic subunits in lignin. Previously characterized bacterial β-etherases are homodimers that fall into two distinct GST subclasses: LigE homologues, which cleave the β(R) stereoisomer of the bond; and LigF homologues, which cleave the β(S) stereoisomer. Here, Great Lakes Bioenergy Research Center researchers report on the discovery of a heterodimeric β-etherase (BaeAB) from the sphingomonad Novosphingobium aromaticivorans that stereospecifically cleaves the β(R)-aryl ether bond of the di-aromatic compound β-(2-methoxyphenoxy)-γ-hydroxypropiovanillone (MPHPV). BaeAB’s subunits are phylogenetically distinct from each other and from other β-etherases, though they are evolutionarily related to LigF, despite the fact that BaeAB and LigF cleave different β-aryl ether bond stereoisomers. The researchers identify amino acid residues in BaeAB’s BaeA subunit important for substrate binding and catalysis, including an asparagine that is proposed to activate the glutathione cofactor. The team also demonstrates that BaeAB homologues from other sphingomonads can cleave β(R)-MPHPV, and that these emzymes may be as common in bacteria as LigE homologues. These results suggest that the ability to cleave the β-aryl ether bond arose independently at least twice in GSTs, and that BaeAB homologues may be important for cleaving the β(R)-aryl ether bonds of lignin-derived oligomers in nature.
Program Manager
N. Kent Peters
kent.peters@science.doe.gov, 301-903-5549
Principal Investigator
Timothy J. Donohue
tdonohue@bact.wisc.edu
Funding
This work was supported by U.S. Department of Energy (DOE) Great Lakes Bioenergy Research Center grants (DOE Office of Science BER DE-FC02-07ER64494 and DE-SC0018409).
Publications
Kontur, W. S. et al. “A heterodimeric glutathione S-transferase that stereospecifically breaks lignin’s β(R)-aryl ether bond reveals the diversity of bacterial β-etherases.” Journal of Biological Chemistry 294, 1877-1890 (2019) [DOI: 10.1074/jbc.RA118.006548].
Related Links
http://www.jbc.org/content/294/6/1877.full