Auto-MuRCiS: a streamlined software package for analysis of multiplex, randomized CRISPR interference sequencing data
K.S. Myers et al. "Auto-MuRCiS: a streamlined software package for analysis of multiplex, randomized CRISPR interference sequencing data" Microbiology Resource Announcements 13:e00356-24 [DOI:10.1128/mra.00356-24]
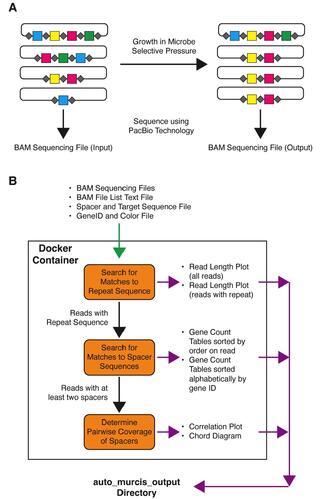
Multiplex, randomized CRISPR interference sequencing (MuRCiS) allows for the simultaneous identification of multiple gene knockouts that together influence microbial processes. Here, we report on an updated analysis tool called Auto-MuRCiS that utilizes Docker to make the analysis of these data rapid and more user-friendly.
The Auto_MuRCiS scripts are available on GitHub: https://github.com/GLBRC/auto_murcis. The Auto_MuRCiS Docker image is available on the Docker Hub (login required): https://hub.docker.com/repository/docker/kevinmyers/auto_murcis/general.