Metagenome study suggests common model of microbial community organization
Background/Objective
Liquid residue from agroindustrial processes are a promising source of valuable chemicals, but it is unclear how to use microbial communities to produce specific products from different residues. Here researchers compared microbial communities started from the same inoculum source and enriched in multiple residues to identify unique and common members.
Approach
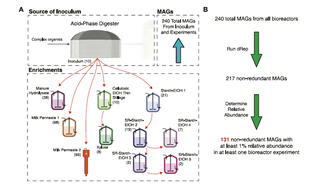
Researchers fed anaerobic bioreactors one of several agroindustrial residues and inoculated them with a microbial community from an acid-phase digester at a wastewater treatment plant. Researchers then identified metagenome-assembled genomes (MAGs) from microbial communities in each bioreactor. To determine if the ecological model was consistent among residues, they used a machine learning algorithm to classify MAGs into three groups: fermenting carbohydrates into intermediate products, using intermediates to produce medium chain fatty acids (MCFAs), and producing MCFAs directly.
Results
Analysis of 217 non-redundant MAGs revealed common biological functions among microbial communities in different bioreactors. While different microorganisms were enriched depending on the agroindustrial residue tested, results supported the conclusion that the microbial ecology model tested was appropriate to explain the chemical production potential from all agricultural residues.
Impact
The study demonstrates the potential of microbial communities to produce chemicals from agroindustrial residues, converting organic waste into useful products and contributing to sustainable circular economy. The machine learning tools can be adapted to evaluate other microbial communities in similar reactor settings.