Machine learning reveals genes impacting oxidative stress resistance across yeasts
Background/Objective
-
Yeasts encounter reactive oxygen species (ROS) during routine metabolism and through interactions with other organisms, including host infections. Scientists have identified key enzymes and genetic regulators in Saccharomyces cerevisiae and a handful of model species but lack deep knowledge about ROS resistance across other species.
Approach
-
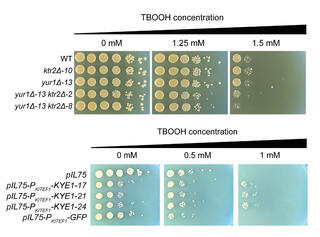
Researchers characterized variation in ROS resistance across the ancient subphylum Saccharomycotina and used machine learning to identify gene families whose sizes were predictive of ROS resistance. The most predictive features were enriched in gene families related to cell wall organization and included two reductase gene families.
Results
-
Experimental validation confirmed that overexpression of the old yellow enzyme reductase increased ROS resistance in Kluyveromyces lactis, while S. cerevisiae mutants lacking multiple mannosyltransferase-encoding genes were hypersensitive to ROS.
Significance/Impacts
-
During industrial overproduction of lipids or proteins, ROS can limit bioproduct yields. Environmental exposure to ROS can lead to the evolution of resistance mechanisms. The study provides a framework for using machine learning to uncover genetic mechanisms that underlie trait variation across diverse species and to inform efforts to genetically engineer desirable traits for clinical and biotechnology applications.
Aranguiz, K., et al., Machine learning reveals genes impacting oxidative stress resistance across yeasts. Nature Communications, 16, 1–15. (2025). [DOI:10.1038/s41467-025-60189-3