Living evidence reveals how thermodynamics shape enzyme investment in glycolysis
Background/Objective
Computational studies predict that thermodynamically constrained reactions and pathways impose greater protein demands on cells, requiring a larger amount of enzyme to sustain a given flux compared to those with stronger thermodynamics. This study provides in vivo evidence that thermodynamic driving forces are a key parameter influencing the enzyme burden of metabolic pathways.
Approach
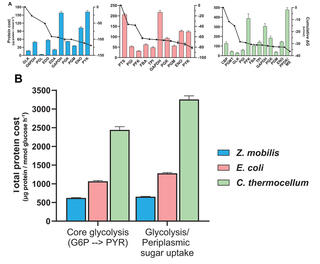
Scientists with the Great Lakes Bioenergy Research Center and the Center for Bioenergy Innovation quantified absolute concentrations of glycolytic enzymes in three bacterial species (Zymomonas mobils, Escherichia coli, and Clostridium thermocellum) that employ distinct glycolytic pathways with varying thermodynamic driving forces and integrated enzyme data with corresponding in vivo metabolic fluxes and ∆G measurements.
Results
Results showed the more thermodynamically favorable Entner-Doudoroff pathway in Z. mobilis requires one fourth as much enzymatic protein to sustain the same flux as the less favorable pryophosphate-dependent glycolytic pathway in C. thermocellum; the Embden-Meyerhof-Parnas pathway in E. coli showed intermediate thermodynamic favorability and enzyme demand. The highly reversible fermentation pathway in C. thermocellum requires 10x more protein than the irreversible pathways in Z. mobilis.
Significance
This study provides in vivo evidence that strongly thermodynamically favorable metabolic pathways require significantly lower enzyme concentrations to sustain a given flux than thermodynamically constrained pathways. The insights and quantitative proteomic data generated will serve as a valuable resource for developing constraint-based genome-scale metabolic models.