Genomic factors shape carbon and nitrogen metabolic niche breadth in yeasts
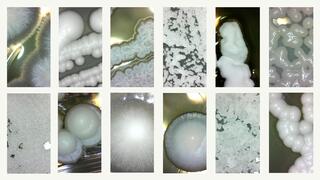
Images of magnified yeast colonies, clockwise from upper left: Alloascoidea africana, Saccharomyces cerevisiae, Peterozyma xylosa, Blastobotrys adeninivorans, Blastobotrys buckinghamii, Lipomyces sp., Sporopachydermia lactativora, Candida boleticola, Hanseniaspora guilliermondii, Ascoidea asiatica, Ambrosiozyma cicatricosa, Candida berthetii
Objective
To explain the extensive variation in ecological niche breadth, from very narrow (specialists) to very broad (generalists).
Approach
Researchers assembled genomic, metabolic, and ecological data for 1,154 yeast strains, which represent nearly all known species in the subphylum Saccharomycotina, and quantified variation in genome sequence, isolation environment, and carbon and nitrogen metabolism.
Results
Analysis showed large interspecific differences in carbon breadth stem from intrinsic differences in genes encoding specific metabolic pathways but no evidence of trade-offs and a limited role of extrinsic ecological factors. The data argue that intrinsic factors shape niche breadth variation in microbes.
Impact
The genomic, metabolic, evolutionary, and ecological data for nearly all known species of the 400-million-year-old yeast subphylum Saccharomycotina provided in this work, coupled with the availability of multiple genetic models in the subphylum, present an inimitable resource and framework for linking genomic variation to phenotypic and ecological variation.