Rewired cellular signaling coordinates sugar and hypoxic responses for anaerobic xylose fermentation in yeast
K.S. Myers et al. (2019). Rewired cellular signaling coordinates sugar and hypoxic responses for anaerobic xylose fermentation in yeast. PLOS Genetics 15: e1008037
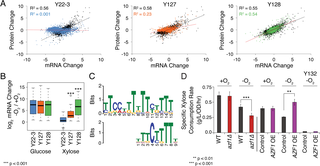
Microbes can be metabolically engineered to produce biofuels and biochemicals, but rerouting metabolic flux toward products is a major hurdle without a systems-level understanding of how cellular flux is controlled. To understand flux rerouting, we investigated a panel of Saccharomyces cerevisiae strains with progressive improvements in anaerobic fermentation of xylose, a sugar abundant in sustainable plant biomass used for biofuel production. We combined comparative transcriptomics, proteomics, and phosphoproteomics with network analysis to understand the physiology of improved anaerobic xylose fermentation. Our results show that upstream regulatory changes produce a suite of physiological effects that collectively impact the phenotype. Evolved strains show an unusual co-activation of Protein Kinase A (PKA) and Snf1, thus combining responses seen during feast on glucose and famine on non-preferred sugars. Surprisingly, these regulatory changes were required to mount the hypoxic response when cells were grown on xylose, revealing a previously unknown connection between sugar source and anaerobic response. Network analysis identified several downstream transcription factors that play a significant, but on their own minor, role in anaerobic xylose fermentation, consistent with the combinatorial effects of small-impact changes. We also discovered that different routes of PKA activation produce distinct phenotypes: deletion of the RAS/PKA inhibitor IRA2 promotes xylose growth and metabolism, whereas deletion of PKA inhibitor BCY1 decouples growth from metabolism to enable robust fermentation without division. Comparing phosphoproteomic changes across ira2Δ and bcy1Δstrains implicated regulatory changes linked to xylose-dependent growth versus metabolism. Together, our results present a picture of the metabolic logic behind anaerobic xylose flux and suggest that widespread cellular remodeling, rather than individual metabolic changes, is an important goal for metabolic engineering.
Raw and processed RNA-seq data were deposited in the NIH GEO database under project number GSE92908. All raw mass spectrometry files and associated information about identifications are available on Chorus under Project ID 999 and Experiment IDs 3007, 3016, and 3166.