Systems-level analysis can improve aromatic funneling by N. aromaticivorans and other bacteria
Background/Objective
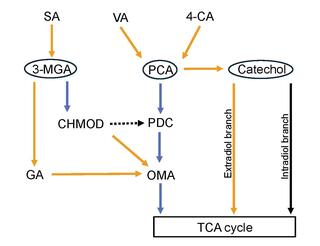
Aromatic compounds are a family of molecules that microbes can metabolize into renewable industrial chemicals. The alphaproteobacterium Novosphingobium aromaticivorans uses multiple pathways to catabolize lignin-associated H-, G- and S-aromatics, but the transcriptional control of the degradation of these molecules is relatively unexplored.
Approach
Researchers performed a systems-level analysis of a LysR-family transcription factor that they named LigR, using DAP-seq, mutant, and in vitro studies to identify and map binding sites within the N. aromaticivorans chromosome. RNA-seq analysis revealed the mode and extent of control of aromatic-inducible genes that depend on LigR.
Results
Cells with deleted LigR were unable to degrade H-, G- and S-aromatics, usually catabolized via protocatechuate meta-cleavage, because of a decreased expression of enzymes involved in this pathway. In vitro assays with the purified transcription factor showed that LigR is a direct activator of these genes. Comparative genomic analysis predicts that LigR homologs and genes directly regulated by this protein in N. aromaticivorans are commonly co-located in Sphingomonadales, but the arrangement is not often found in other known aromatic-metabolizing bacteria.
Significance/Impact
Lignin-derived aromatics represent a renewable source of industrial chemicals. A systems-level understanding aromatic metabolism can inform engineering of commercially relevant bacteria that can produce these chemicals or efforts to remove toxic aromatics from the production process.
Rodríguez-Castro, L., Myers, K. S., Linz, A. M., Mettert, E. L., Camp, W., Kiley, P. J., Noguera, D. R., & Donohue, T. J. Novosphingobium aromaticivorans LigR coordinates transcription of genes involved in metabolism of multiple types of aromatics. mSystems, 0, e01469-25. (2025). [DOI:10.1128/msystems.01469-25]