Independent chemical genomics approaches predict targets for increasing chemical tolerance
Background/Objective
Screens of gene perturbation libraries against biofuel production stressors can identify high-value engineering targets, but follow-up experiments needed to guard against false positives are slow and resource intensive.
Approach
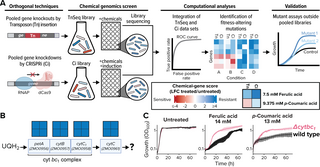
Scientists screened genome-scale CRISPRi (CRISPR interference) and TnSeq (transposon insertion sequencing) libraries of the Alphaproteobacterium Zymomonas mobilis against growth inhibitors commonly found in deconstructed biomass. By integrating the data from the complementary techniques they established an approach for defining engineering targets with high specificity.
Results
The screens identified all known genes in the cytochrome bc1 and cytochrome c synthesis pathway as potential targets for engineering resistance to phenolic acids under anaerobic conditions. Results also showed exposure of Z. mobilis to ferulic acid (a phenolic acid) causes substantial remodeling of the cell envelope proteome as well as downregulation of TonB-dependent transporters.
Impact
This work revealed genetic modifications that improve anaerobic growth of Z. mobilis in the presence of inhibitory chemicals found in plant-based feedstocks. This method can be broadly applied to predict microbial responses to chemicals across systems, paving the way for developments in biomanufacturing, therapeutics, and agriculture.