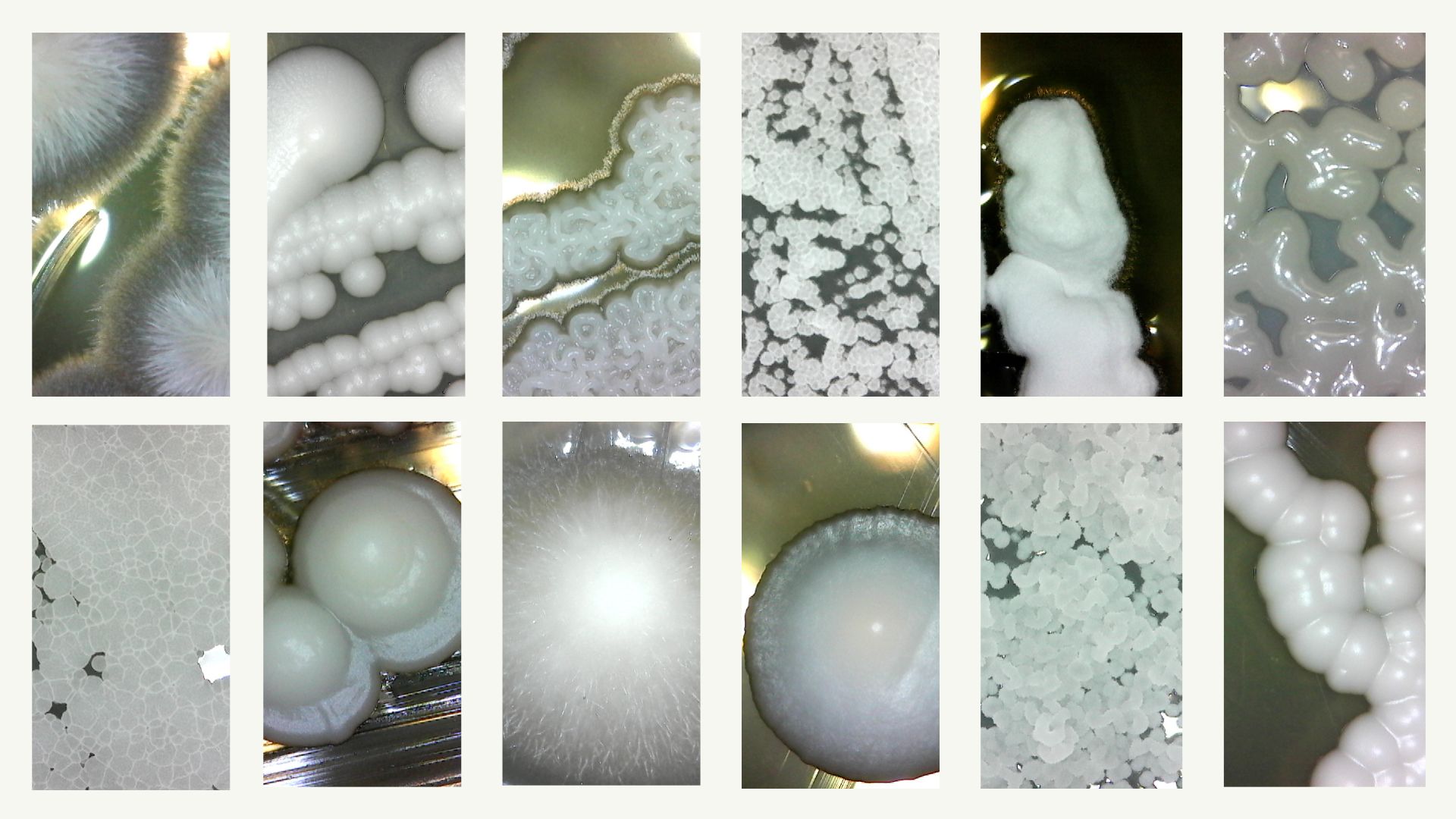
In a landmark study based on one of the most comprehensive genomic datasets ever assembled, a team led by scientists at the University of Wisconsin–Madison and Vanderbilt University offer a possible answer to one of the oldest questions about evolution: why some species are generalists and others specialists.
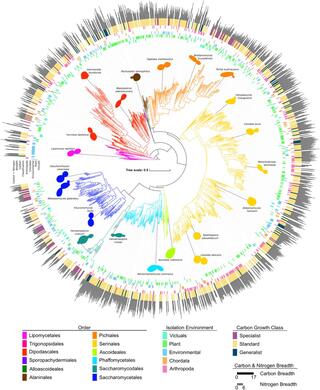
Under the guidance of UW–Madison Professor of Genetics Chris Todd Hittinger and Antonis Rokas, professor of biology at Vanderbilt University, researchers mapped the genetic blueprints, appetites, and environments of more than 1,000 species of yeasts, building a family tree that illuminates how these single-celled fungi evolved over the past 400 million years.
The results, published April 25 in the journal Science, suggest that internal – not external – factors are the primary drivers of variation in the types of carbon yeasts can eat, and the researchers found no evidence that metabolic versatility, or the ability to eat different foods, comes with any trade-offs. In other words, some yeasts are jacks-of-all-trades and masters of each.
That really, really surprised us,” Hittinger said, “Specialists should be better at the carbon sources for which they are specialized. And generalists, if they’re eating everything, they should not be as good. And instead that’s not what we see.”
The paper is a product of an ongoing decade-long project to build a comprehensive database mapping the relationship between genomes and traits of yeasts, a group of species as genetically diverse as all animals. The genomic dataset is the most comprehensive ever compiled for such an ancient and diverse group.
Hittinger, an investigator with the Great Lakes Bioenergy Research Center who studies yeast metabolism, said in addition to furthering our understanding of biodiversity, the database can help researchers identify or create yeasts that are better at converting plant sugars into biofuels and other alternatives to fossil fuels.
Many branches, varied appetites
Starting in 2015, Hittinger’s team sequenced the genomes and studied the metabolisms of nearly every known species of a group of yeasts distantly related to Saccharomyces cerevisiae, better known as baker’s yeast.
They chose this group because of the wide array of species that had been identified and their highly variable carbon diets.

“We have lots of branches, some that are close together, some that are further apart,” Hittinger said. “You just have tons of opportunity for the same or similar evolutionary trajectories to be explored. We can see traits that have been gained or lost a dozen times.”
What they didn’t know is how the species were related.
After assembling the data, researchers used machine learning tools to figure out which genes are associated with which traits, including the range of resources an organism can use or the conditions it can tolerate — a concept known as "niche breadth."
Like other organisms, some yeasts have evolved to be specialists — think koalas, which eat nothing but eucalyptus leaves – while others are generalists like raccoons, which eat just about anything.
Scientists have been trying to explain why both generalists and specialists exist almost since Charles Darwin proposed his theory of evolution in 1859.
“Those ideas were percolating in the time of Darwin, and soon thereafter, as people started to … hone in on ecology as the basis of how natural selection works,” Hittinger said.
Scientists have offered two broad models to explain the phenomenon.
One suggests generalists are jacks-of-all-trades but masters of none, meaning they can tolerate a wider range of conditions or food sources but aren’t as dominant as a specialist in any specific niche.
The other theory is that a combination of internal and external factors drive niche variation.
For example, organisms can acquire genes that allow them to make enzymes capable of breaking down more than one substance, expanding the range of foods they can eat. Conversely, random loss of genes over time can result in a narrower palate.
Likewise, environments can exert selective pressure on traits. So a habitat with only one or two food sources or constant temperatures would favor specialists, while generalists might do better in an environment with a wider array of food or conditions.

When it comes to yeast metabolism, Hittinger’s team found no evidence of trade-offs.
“The generalists are better across all the carbon sources they can use,” Hittinger said. “Generalists are also able to use more nitrogen sources than carbon specialists. I wouldn't have predicted that relationship at all.”
The data also showed that environmental factors play only a limited role.
That too was surprising, said co-author Dana Opulente, who began the project as a postdoctoral researcher at UW–Madison and is now a professor of biology at Villanova University.
“We might expect to find specialists mostly in domesticated strains, but that's not the case,” Opulente said. “We can find generalists and specialists in soil and on flowers. We’re finding them in all the same places.”
Hittinger cautions there are limitations to what can be inferred from the data. It’s possible that tradeoffs are present in species that weren’t studied. And the lab experiments used to measure metabolic growth can’t replicate the conditions in soils, tree bark, or insect guts where yeasts live in nature.

Opulente is now working to gather more data on those natural environments, which could reveal a stronger ecological influence on niche breadth.
“If we have more data, there's a lot of other questions that could be asked,” Opulente said.
The study also does not explain why, if there are no tradeoffs, all yeasts aren’t generalists.
One possible explanation is that genes often disappear during evolution, and so long as it isn’t essential for survival that mutation can get passed on and take over a population. Specialists might continually evolve from generalists through this process.
“I'm not sure that we’ve answered that question yet,” Hittinger said.
Putting yeasts to work
Like humans and animals, yeasts can’t make their own food. Instead they rely on carbon that plants have stored as energy sources such as sugars.
“They vary a lot in terms of what they can eat,” Hittinger said. “They also make very different decisions about what they're going to do with that carbon.”
For example, most yeasts will use oxygen when it’s available to convert sugars into carbon dioxide and water, capturing energy in the process. But Saccharomyces cerevisiae only uses some of the carbon, converting most of it into ethanol.
Energetically it’s a wasteful strategy, but leaving food on the plate is what makes this yeast a workhorse of the food, beverage, and biofuels industries.
Hittinger hopes the database will help identify other useful species or new genetic pathways to get around biological obstacles, such as tricking yeasts into eating xylose when they prefer glucose.
“It is helping point to species and lineages that have unique potentials that have not yet been exploited,” Hittinger said. “But maybe even more interesting and informative … is that by having the same thing happen 5, 10, 15 times, that gives us more power to identify the genetic parts and specific changes that have happened during evolution.”
The project was funded by a $2 million grant from the National Science Foundation’s Dimensions of Biodiversity program. With the help of a second NSF grant, Hittinger’s team and collaborators at Vanderbilt University are now working to build a more detailed genotype-phenotype map.
“Now we get to dig in and go deeper and say, Okay, we know it’s these genes and we know it’s this phenotype,” Opulente said. “But how does that work?”
This work was supported by grants from the National Science Foundation; the Office of Science, U.S. Department of Energy; U.S. Department of Agriculture; the Wisconsin Alumni Research Foundation; the National Institutes of Health and National Institute of Allergy and Infectious Diseases; the Burroughs Wellcome Fund; the National Key R&D Program of China, National Science Foundation for Distinguished Young Scholars of Zhejiang Province; Fundamental Research Funds for the Central Universities of China; the Howard Hughes Medical Institute; the Slovenian Research Agency; Foundation for Science and Technology (Portugal); National Council for Scientific and Technological Development (Brazil); National Scientific and Technical Research Council (CONICET), Argentina; the National University of Comahue (Argentina).