The Science
Reactive oxygen species (ROS) are unstable molecules created during natural functions like breathing and converting food into energy. During industrial processes, yeasts are often exposed to high levels of these oxidants, which can limit how much high-value fatty acids or other bioproducts they can make. Some yeasts are naturally resistant to oxidative stress, but scientists have limited knowledge of which species or how they protect themselves. Here, scientists used machine learning to analyze the genetic blueprints of hundreds of yeasts and identify which groups of genes are most important for ROS resistance.
The Impact
Identifying the genetic factors related to oxidative stress resistance can help scientists understand how these functions evolved. The study highlights genes that could be targeted to make more robust strains for industrial uses, like making biofuels or bioproducts, or to fight yeast infections. The work also provides a framework for using machine learning to identify genes related to traits across many species.
Summary
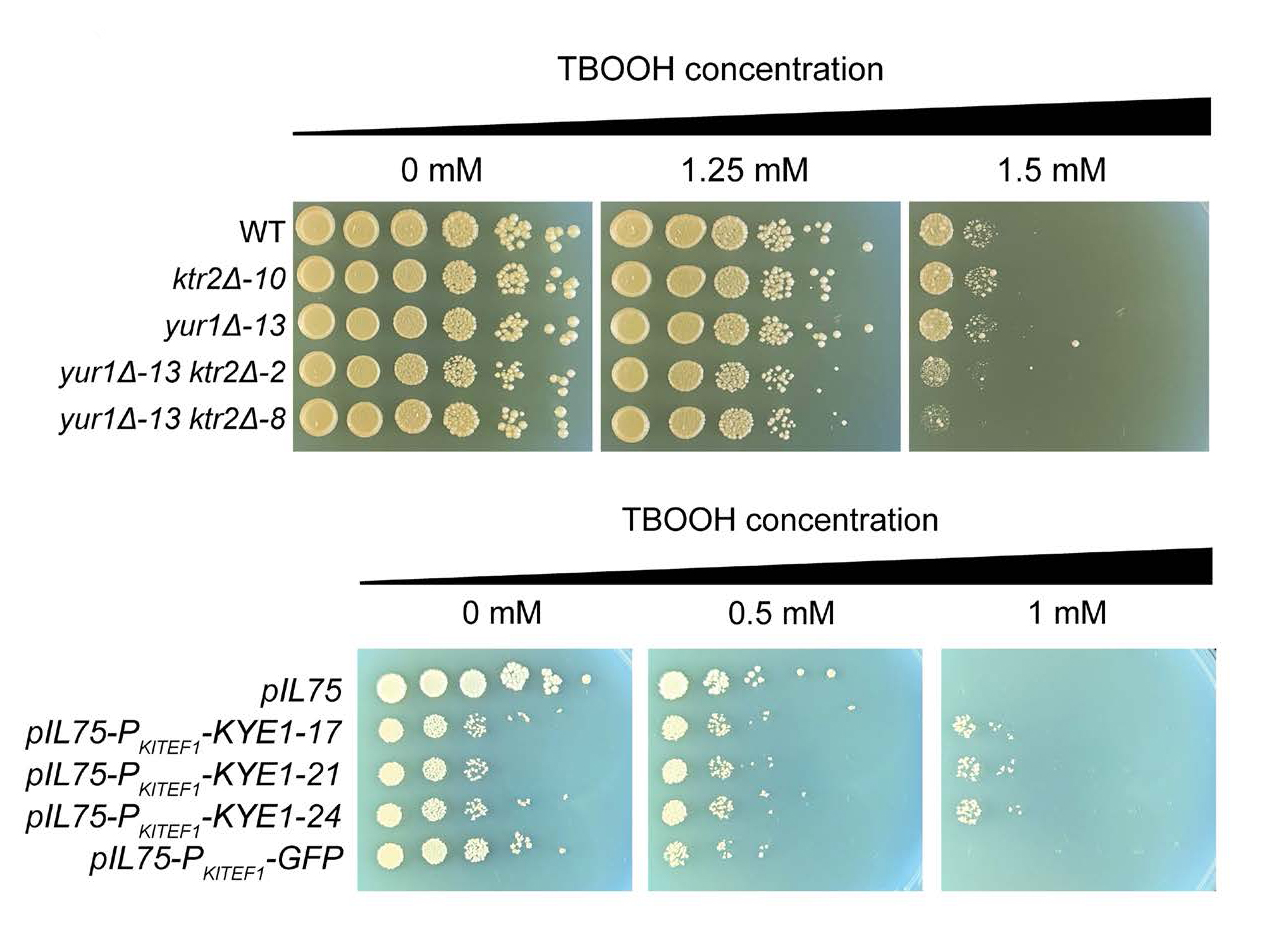
Scientists with the Great Lakes Bioenergy Research Center tested 285 yeasts across the subphylum Saccharomycotina to evaluate the variance in ROS resistance. They then trained a machine learning model to identify the genes most important for resistance. The model identified two groups of important genes: one helps build and maintain the cell wall; the others are reductase genes that produce an enzyme to neutralize ROS.
To validate the predictions, the scientists did two experiments. First, they took a species highly susceptible to ROS, Kluyveromyces lactis, and gave it an extra copy of the reductase gene to increase production of the “old yellow enzyme.” The modified yeast was more resistant to ROS. Researchers then deleted two genes responsible for cell wall construction from a second species, Saccharomyces cerevisiae, which made that yeast more susceptible to ROS stress. These experiments show that we can draw connections between machine learning model predictions and benchtop biology.