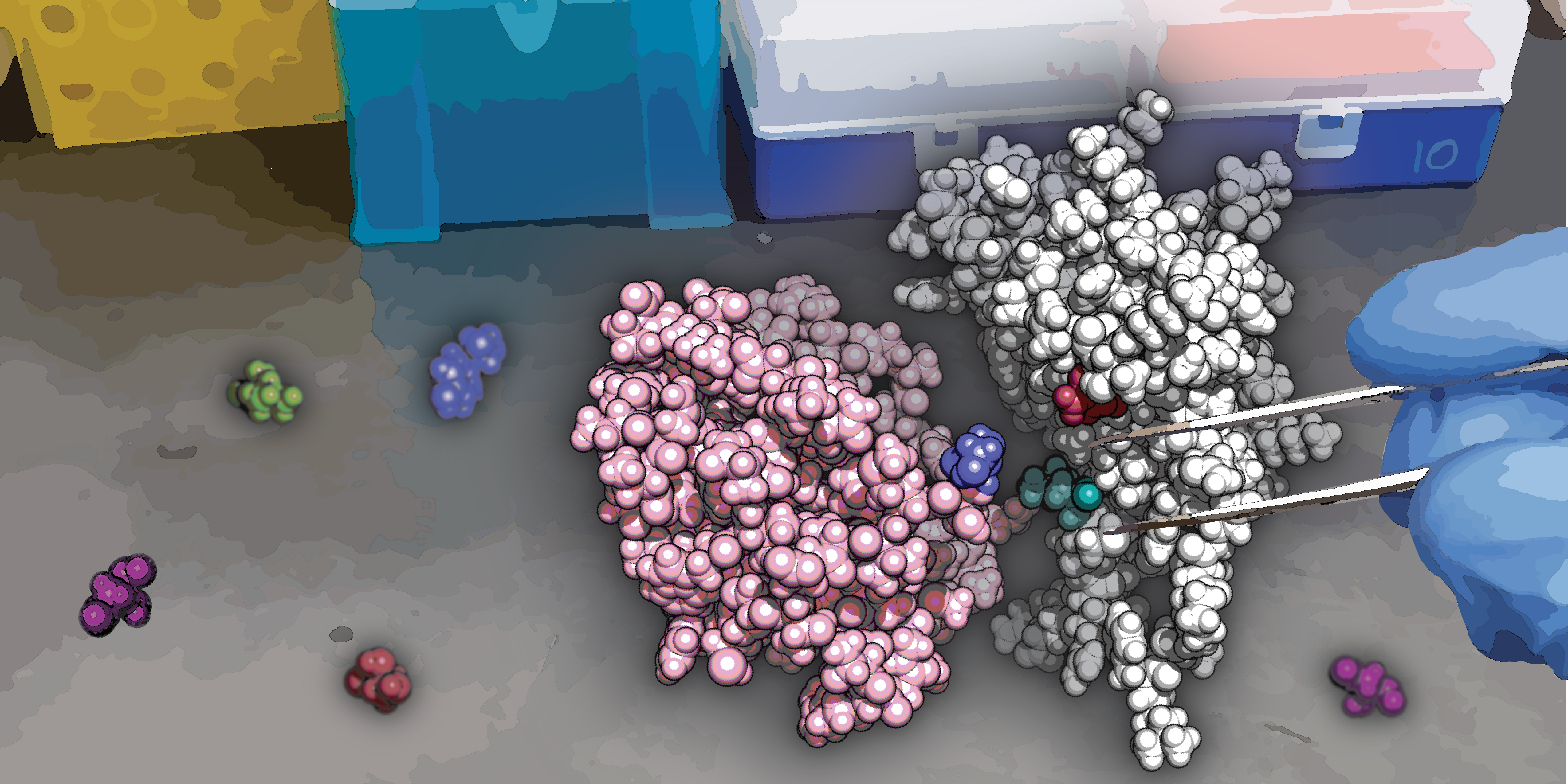
Split protein systems are biochemical tools that can be used to monitor and regulate biological activity. A protein of interest is broken into two inactive pieces that, under user-defined conditions, can rejoin to form a functional protein. However, it can be difficult to engineer a split such that the protein will reconstitute effectively only under the desired conditions.
In a paper published Feb. 1, 2021, in Nature Chemical Biology, Great Lakes Bioenergy Research Center (GLBRC) researcher and University of Wisconsin–Madison biochemistry assistant professor Vatsan Raman, graduate student Anthony Meger, and colleagues at Northwestern University describe a computation-guided method, called Split Protein Optimization by Reconstitution Tuning (SPORT), to optimize the design of split protein systems. Here, Raman describes the challenges involved in splitting proteins, how SPORT helps solve them, and potential uses for bioenergy researchers.

How do split protein systems work and why are they useful?
Proteins carry out their biological function as intact biomolecules. Any effort to truncate or cut proteins makes them structurally unstable and incapable of carrying out their function. However, a protein can be carefully split without completely destabilizing it, such that each half is structurally stable, but the protein is nonfunctional. But under certain conditions—which you as a user decide—the halves can come back together and make the protein functional again. This gives a researcher full control over the protein function and, by extension, the cellular processes mediated by that protein. Split proteins are a powerful biotechnological tool to understand and control cellular functions.
A simple example is splitting green fluorescent protein, GFP. Say you split GFP and connect the halves to two proteins, A and B. Only when A and B interact will the split halves come together and GFP light up. If you want to know under what conditions A and B interact, you only have to monitor fluorescence from GFP. That makes the experiment simple, fast, and scalable to many conditions. Or, instead of GFP, you could split an enzyme. For instance, if you want a certain enzyme to be active only when two cell receptors are in close proximity, then each split half of the enzyme can be attached to a different receptor.
What are some existing challenges with using these systems, and how will SPORT help?
Nature has not evolved proteins to be split. Think about the architecture of a protein: the central core of the protein is mostly sticky residues—hydrophobic amino acids—and toward the periphery you’ll often find charged or polar amino acids. If you split a protein naïvely, the two halves will just stick back—a Velcro-like effect—because the hydrophobic amino acids prefer being around each other. To make a conditionally active split protein system, you have to mutate that interface so that the protein comes back together only under the specific conditions that you decide on, not by itself.
That turns out to be challenging, because when one mutates the central core of a protein, the protein becomes highly unstable. So, a protein designer has to walk this tightrope between preventing the protein halves from rejoining by that Velcro-like effect, and destabilizing the protein altogether. Those are the two extremes. SPORT computationally estimates the “Goldilocks zone” where the protein can be split without spontaneous association (Velcro) and without destabilization. In thermodynamic terms, it finds the energy window between spontaneous reconstitution and complete disassembly. One can make mutations that fall within that energy window, and you get this nice conditionally active split protein activity.
What do you see as some of the most exciting potential uses of split-protein systems in bioenergy research?
An important application area that I see for the GLBRC is in conversion, where the goal is to take sugars and lignin from degraded plants and feed them to microbes to make valuable products. Microbial metabolic pathways for converting sugars to valuable products involve many enzymes, and often some enzymes need to be turned off and turned on under certain conditions. This tool would be useful in multi-enzyme metabolic engineering, where we seek to exert some degree of control over the pathway. One example might be avoiding the of accumulation of toxic byproducts. Another could be if you want to make one product until a certain point and then switch to a different product.
How accessible is this design-driven strategy for researchers? Does it take specialized knowledge or equipment?
Our broad goal is to make split proteins on demand. In other words, you give us an enzyme that you're interested in splitting, and we’ll recommend mutations that will make your conditionally active split protein system. Running SPORT calculations is quite straightforward. But there may be a barrier to entry if one isn’t familiar with computational molecular modeling. We are happy to work with labs in GLBRC or other bioenergy research centers to run SPORT for their protein of interest. We can provide some mutants that we predict will have desired properties, and they will have to experimentally measure the activity of our mutants. Eventually, we would like to have a web server where researchers can upload their protein of interest and the webserver returns mutations to test.
How are you using SPORT in your own research?
We've only just developed SPORT, so we haven't really applied it to anything yet except for the viral protease that we published in the paper. We have started a collaboration to test other candidates. But we are also on the lookout for new cases to test SPORT. Most enzymes are good targets because they are nice globular structures, which will make them amenable to SPORT. We are looking for candidate proteins, and we welcome collaborations from GLBRC researchers.
Read the paper
Dolberg, T. B., et al. Computation-guided optimization of split protein systems. Nature Chemical Biology. Published online February 1, 2021. https://doi.org/10.1038/s41589-020-00729-8