Widespread irreproducibility in phylogenetics
X. Shen et al. “Widespread irreproducibility in phylogenetics” Nature Communications 11, 6096 (2020) [DOI: 10.1038/s41467-020-20005-6]
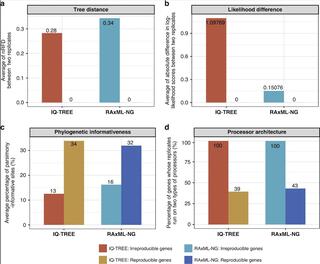
Phylogenetic trees are essential for studying biology, but their reproducibility under identical parameter settings remains unexplored. Here, we find that 3515 (18.11%) IQ-TREE-inferred and 1813 (9.34%) RAxML-NG-inferred maximum likelihood (ML) gene trees are topologically irreproducible when executing two replicates (Run1 and Run2) for each of 19,414 gene alignments in 15 animal, plant, and fungal phylogenomic datasets. Notably, coalescent-based ASTRAL species phylogenies inferred from Run1 and Run2 sets of individual gene trees are topologically irreproducible for 9/15 phylogenomic datasets, whereas concatenation-based phylogenies inferred twice from the same supermatrix are reproducible. Our simulations further show that irreproducible phylogenies are more likely to be incorrect than reproducible phylogenies. These results suggest that a considerable fraction of single-gene ML trees may be irreproducible. Increasing reproducibility in ML inference will benefit from providing analyses’ log files, which contain typically reported parameters (e.g., program, substitution model, number of tree searches) but also typically unreported ones (e.g., random starting seed number, number of threads, processor type).