Transcriptional regulation of the raffinose family oligosaccharides pathway in Sorghum bicolor reveals potential roles in leaf sucrose transport and stem sucrose accumulation
B.A. McKinley et al. "Transcriptional regulation of the raffinose family oligosaccharides pathway in Sorghum bicolor reveals potential roles in leaf sucrose transport and stem sucrose accumulation" Frontiers in Plant Science 12 (2022) [DOI: 10.3389/fpls.2022.1062264]
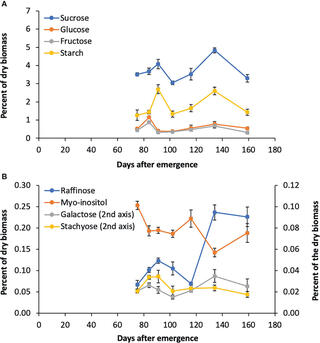
Bioenergy sorghum hybrids are being developed with enhanced drought tolerance and high levels of stem sugars. Raffinose family oligosaccharides (RFOs) contribute to plant environmental stress tolerance, sugar storage, transport, and signaling. To better understand the role of RFOs in sorghum, genes involved in myo-inositol and RFO metabolism were identified and relative transcript abundance analyzed during development. Genes involved in RFO biosynthesis (SbMIPS1, SbInsPase, SbGolS1, SbRS) were more highly expressed in leaves compared to stems and roots, with peak expression early in the morning in leaves. SbGolS, SbRS, SbAGA1 and SbAGA2 were also expressed at high levels in the leaf collar and leaf sheath. In leaf blades, genes involved in myo-inositol biosynthesis (SbMIPS1, SbInsPase) were expressed in bundle sheath cells, whereas genes involved in galactinol and raffinose synthesis (SbGolS1, SbRS) were expressed in mesophyll cells. Furthermore, SbAGA1 and SbAGA2, genes that encode neutral-alkaline alpha-galactosidases that hydrolyze raffinose, were differentially expressed in minor vein bundle sheath cells and major vein and mid-rib vascular and xylem parenchyma. This suggests that raffinose synthesized from sucrose and galactinol in mesophyll cells diffuses into vascular bundles where hydrolysis releases sucrose for long distance phloem transport. Increased expression (>20-fold) of SbAGA1 and SbAGA2 in stem storage pith parenchyma of sweet sorghum between floral initiation and grain maturity, and higher expression in sweet sorghum compared to grain sorghum, indicates these genes may play a key role in non-structural carbohydrate accumulation in stems.
Data from this study are hosted by the JGI Genome Portal: 1. Sorghum bicolor Della internode development, JGI Project ID: 1190869. 2. Sorghum bicolor Della leaves development, JGI Project ID: 1240803. 3. Sorghum bicolor Circadian Cycling gene expression profiling, JGI Project ID: 1051031. 4. Sorghum bicolor BTx623 Gene Atlas (plate 8, 9, and 10), JGI Project IDs: 1051038, 1051407, 1053826.