Transcriptional competition shapes proteotoxic ER stress resolution
D. Kwan Ko and F. Brandizzi "Transcriptional competition shapes proteotoxic ER stress resolution" Nature Plants 8 (2022) [DOI: 10.1038/s41477-022-01150-w]
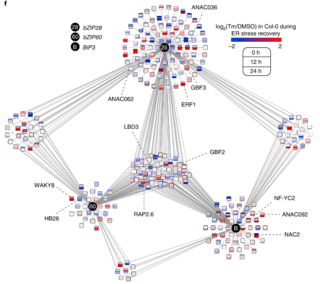
Through dynamic activities of conserved master transcription factors (mTFs), the unfolded protein response (UPR) relieves proteostasis imbalance of the endoplasmic reticulum (ER), a condition known as ER stress. Because dysregulated UPR is lethal, the competence for fate changes of the UPR mTFs must be tightly controlled. However, the molecular mechanisms underlying regulatory dynamics of mTFs remain largely elusive. Here, we identified the abscisic acid-related regulator G-class bZIP TF2 (GBF2) and the cis-regulatory element G-box as regulatory components of the plant UPR led by the mTFs, bZIP28 and bZIP60. We demonstrate that, by competing with the mTFs at G-box, GBF2 represses UPR gene expression. Conversely, a gbf2 null mutation enhances UPR gene expression and suppresses the lethality of a bzip28 bzip60 mutant in unresolved ER stress. By demonstrating that GBF2 functions as a transcriptional repressor of the UPR, we address the long-standing challenge of identifying shared signalling components for a better understanding of the dynamic nature and complexity of stress biology. Furthermore, our results identify a new layer of UPR gene regulation hinged upon an antagonistic mTFs-GFB2 competition for proteostasis and cell fate determination.
The ChIP–seq data supporting the finding of this study have been deposited in the NCBI Sequence Read Archive and are accessible through the BioProject accession code PRJNA810750. The scripts used in this study are available on GitHub.