Stable hypermutators revealed by the genomic landscape of genes involved in genome stability among yeast species
C. Gonçalves et al. "Stable hypermutators revealed by the genomic landscape of genes involved in genome stability among yeast species" Molecular Biology and Evolution (2025) 42:msaf285 [DOI:10.1093/molbev/msaf285]
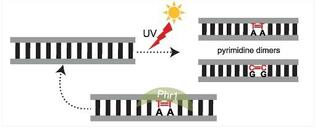
Mutator phenotypes are short-lived due to the rapid accumulation of deleterious mutations. Yet, recent observations reveal that certain fungi can undergo prolonged accelerated evolution after losing genes involved in DNA repair. Here, we surveyed 1,154 yeast genomes representing nearly all known yeast species of the subphylum Saccharomycotina (phylum Ascomycota) to examine the relationship between reduced gene repertoires broadly associated with genome stability functions (e.g., DNA repair, cell cycle) and elevated evolutionary rates. We identified three distantly related lineages—encompassing 12% of species—that had both the most streamlined sets of genes involved in genome stability (specifically DNA repair) and the highest evolutionary rates in the entire subphylum. Two of these “faster-evolving lineages” (FELs)—a subclade within the order Pichiales and the Wickerhamiella/Starmerella (W/S) clade (order Dipodascales)—are described here for the first time, while the third corresponds to a previously documented Hanseniaspora FEL. Examination of genome stability gene repertoires revealed a set of genes predominantly absent in these three FELs, suggesting a potential role in the observed acceleration of evolutionary rates. In the W/S clade, genomic signatures are consistent with a substantial mutational burden, including pronounced A|T bias and endogenous DNA damage. Interestingly, we found that the W/S clade also contains DNA repair genes possibly acquired through horizontal gene transfer, including a photolyase of bacterial origin. These findings highlight how hypermutators can persist across macroevolutionary timescales, potentially linked to the loss of genes related with genome stability, with horizontal gene transfer as a possible avenue for partial functional compensation.
All input data for all analyses and raw results files can be found in Figshare: https://figshare.com/s/3d575d6283c970a0c1e4. Genomic data used in this work is publicly available and can be accessed via the Figshare repository from Opulente et al. (2024): https://plus.figshare.com/articles/dataset/Genomes_and_Annotations_of_1_154_budding_yeasts/22802147.