Rapid Targeted Quantitation of Protein Overexpression with Direct Infusion Shotgun Proteome Analysis (DISPA-PRM)
E.A. Trujillo et al. "Rapid Targeted Quantitation of Protein Overexpression with Direct Infusion Shotgun Proteome Analysis (DISPA-PRM)" Analytical Chemistry 94 (2022) [DOI: 10.1021/acs.analchem.1c03243]
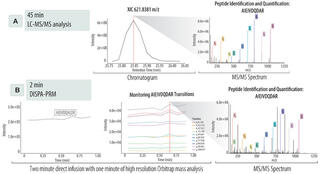
While much effort has been placed on comprehensive quantitative proteome analysis, certain applications demand the measurement of only a few target proteins from complex systems. Traditional approaches to targeted proteomics rely on nanoliquid chromatography (nLC) and targeted mass spectrometry (MS) methods, e.g., parallel reaction monitoring (PRM). However, the time requirement for nLC can limit the throughput of targeted proteomics. To achieve rapid and high-throughput targeted methods, here we show that nLC separations can be eliminated and replaced with direct infusion shotgun proteome analysis (DISPA) using high-field asymmetric waveform ion mobility spectrometry (FAIMS) with PRM. We demonstrate the application of DISPA-PRM for rapid targeted quantification of bacterial enzymes utilized in the production of biofuels by monitoring temporal expression in 72 metabolically engineered bacterial cultures in less than 2.5 h, with a measured dynamic range >1200-fold. We conclude that DISPA-PRM presents a valuable innovative tool with results comparable to nLC-MS/MS, enabling fast and rapid detection of targeted proteins in complex mixtures.
Protein and peptide metrics from nLC-MS/MS can be found in Supporting Information. The codes used in the study can be found on GitHub.