Population genomics of the pathogenic yeast Candida tropicalis identifies hybrid isolates in environmental samples
C.E. O’Brien et al. "Population genomics of the pathogenic yeast Candida tropicalis identifies hybrid isolates in environmental samples" PLoS Pathogens 17, 3 (2021) [DOI: 10.1371/journal.ppat.1009138]
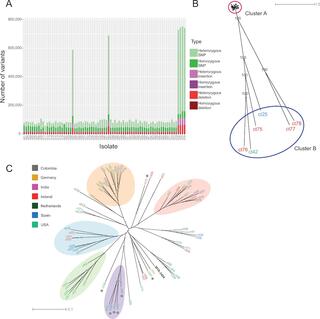
Candida tropicalis is an important fungal pathogen, which is particularly common in the Asia-Pacific and Latin America. There is currently very little known about the diversity of genotype and phenotype of C. tropicalis isolates. By carrying out a phylogenomic analysis of 77 isolates, we find that C. tropicalis genomes range from very homozygous to highly heterozygous. We show that the heterozygous isolates are hybrids, most likely formed by mating between different parents. Unlike other Candida species, the hybrids are more common in environmental than in clinical niches, suggesting that for this species, hybridization is not associated with virulence. We also explore the range of phenotypes, and we identify a genomic variant that is required for growth on valine and isoleucine as sole nitrogen sources.
All sequencing data is available at NCBI under BioProject accession PRJNA604451 and C. tropicalis genome assembly B and annotation is available under NCBI accession JAFIQD000000000. rDNA sequences are available at accession numbers MW584905- MW584910. Other data sets (i.e. variant calls and images for phenotype analysis) are available in Figshare.