Ploidy evolution in a wild yeast is linked to an interaction between cell type and metabolism
J.G. Crandall et al. "Ploidy evolution in a wild yeast is linked to an interaction between cell type and metabolism" PLOS Biology 21:e3001909 (2023) [DOI:10.1371/journal.pbio.3001909]
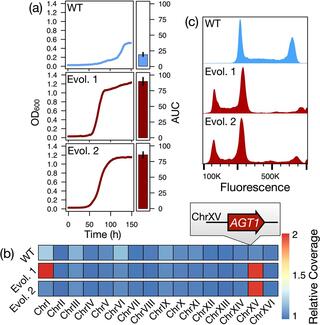
Ploidy is an evolutionarily labile trait, and its variation across the tree of life has profound impacts on evolutionary trajectories and life histories. The immediate consequences and molecular causes of ploidy variation on organismal fitness are frequently less clear, although extreme mating type skews in some fungi hint at links between cell type and adaptive traits. Here, we report an unusual recurrent ploidy reduction in replicate populations of the budding yeast Saccharomyces eubayanus experimentally evolved for improvement of a key metabolic trait, the ability to use maltose as a carbon source. We find that haploids have a substantial, but conditional, fitness advantage in the absence of other genetic variation. Using engineered genotypes that decouple the effects of ploidy and cell type, we show that increased fitness is primarily due to the distinct transcriptional program deployed by haploid-like cell types, with a significant but smaller contribution from absolute ploidy. The link between cell-type specification and the carbon metabolism adaptation can be traced to the noncanonical regulation of a maltose transporter by a haploid-specific gene. This study provides novel mechanistic insight into the molecular basis of an environment-cell type fitness interaction and illustrates how selection on traits unexpectedly linked to ploidy states or cell types can drive karyotypic evolution in fungi.
Strains and plasmids are available upon request. All raw sequencing data has been deposited at NCBI SRA under BioProject PRJNA894214.