Phyllosphere exudates select for distinct microbiome members in sorghum epicuticular wax and aerial root mucilage
M. Mechan et al. "Phyllosphere exudates select for distinct microbiome members in sorghum epicuticular wax and aerial root mucilage" Phytobiomes (2023) [DOI:10.1094/PBIOMES-08-22-0046-FI]
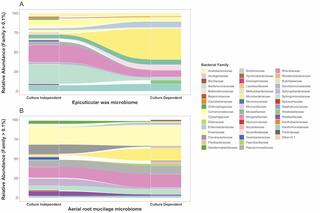
Phyllosphere exudates create specialized microhabitats that shape microbial community diversity. We explored the microbiome associated with two sorghum phyllosphere exudates, the epicuticular wax and aerial root mucilage. We assessed the microbiome associated with the wax from sorghum plants over two growth stages, and the root mucilage additionally from nitrogen-fertilized and non-fertilized plants. In parallel, we isolated and characterized hundreds of bacteria from wax and mucilage, and integrated data from cultivation-independent and cultivation-dependent approaches to gain insights into exudate diversity and bacterial phenotypes. We found that Sphingomonadaceae and Rhizobiaceae families were the major taxa in the wax regardless of water availability and plant developmental stage to plants. The cultivation-independent mucilage-associated bacterial microbiome contained Erwiniaceae, Flavobacteriaceae, Rhizobiaceae, Pseudomonadaceae, Sphingomonadaceae, and its structure was strongly influenced by sorghum development but only modestly influenced by fertilization. In contrast, the fungal community structure of mucilage was strongly affected by the year of sampling but not by fertilization or plant developmental stage, suggesting a decoupling of fungal-bacterial dynamics in the mucilage. Our bacterial isolate collection from wax and mucilage had several isolates that matched 100% to detected amplicon sequence variants, and were enriched on media that selected for phenotypes including phosphate solubilization, putative diazotrophy, resistance to desiccation, capability to grow on methanol as a carbon source, and ability to grow in the presence of linalool and β-caryophyllene (terpenes in sorghum wax). This work expands our understanding of the microbiome of phyllosphere exudates and supports our long-term goal to translate microbiome research to support sorghum cultivation.
The data analysis workflows for sequence processing and ecological statistics are available on GitHub. Raw sequencing data has been deposited in the Sequence Read Archive NCBI database under BioProject accession number PRJNA844896. Full-length 16S rRNA sequence data has been deposited in the GenBank with accession numbers ON973084-ON973283.