Oxygenation influences xylose fermentation and gene expression in the yeast genera Spathaspora and Scheffersomyces
K.O. Barros et al. "Oxygenation influences xylose fermentation and gene expression in the yeast genera Spathaspora and Scheffersomyces" Biotechnology for Biofuels and Bioproducts 17:20 (2024) [DOI:10.1186/s13068-024-02467-8]
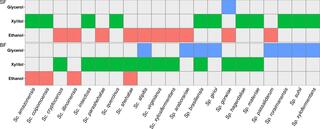
BACKGROUND: Cost-effective production of biofuels from lignocellulose requires the fermentation of D-xylose. Many yeast species within and closely related to the genera Spathaspora and Scheffersomyces (both of the order Serinales) natively assimilate and ferment xylose. Other species consume xylose inefficiently, leading to extracellular accumulation of xylitol. Xylitol excretion is thought to be due to the different cofactor requirements of the first two steps of xylose metabolism. Xylose reductase (XR) generally uses NADPH to reduce xylose to xylitol, while xylitol dehydrogenase (XDH) generally uses NAD(+) to oxidize xylitol to xylulose, creating an imbalanced redox pathway. This imbalance is thought to be particularly consequential in hypoxic or anoxic environments. RESULTS: We screened the growth of xylose-fermenting yeast species in high and moderate aeration and identified both ethanol producers and xylitol producers. Selected species were further characterized for their XR and XDH cofactor preferences by enzyme assays and gene expression patterns by RNA-Seq. Our data revealed that xylose metabolism is more redox balanced in some species, but it is strongly affected by oxygen levels. Under high aeration, most species switched from ethanol production to xylitol accumulation, despite the availability of ample oxygen to accept electrons from NADH. This switch was followed by decreases in enzyme activity and the expression of genes related to xylose metabolism, suggesting that bottlenecks in xylose fermentation are not always due to cofactor preferences. Finally, we expressed XYL genes from multiple Scheffersomyces species in a strain of Saccharomyces cerevisiae. Recombinant S. cerevisiae expressing XYL1 from Scheffersomyces xylosifermentans, which encodes an XR without a cofactor preference, showed improved anaerobic growth on xylose as the primary carbon source compared to S. cerevisiae strain expressing XYL genes from Scheffersomyces stipitis. CONCLUSION: Collectively, our data do not support the hypothesis that xylitol accumulation occurs primarily due to differences in cofactor preferences between xylose reductase and xylitol dehydrogenase; instead, gene expression plays a major role in response to oxygen levels. We have also identified the yeast Sc. xylosifermentans as a potential source for genes that can be engineered into S. cerevisiae to improve xylose fermentation and biofuel production.
Supporting data are available in additional files. Raw RNA-Seq reads of Sc. xylosifermentans CBS 12540T, Sc. coipomoensis NRRL Y-17651T, and Sc. amazonensis UFMG-HMD-26.3T are available on the JGI portal http://genome.jgi.doe.gov and have been deposited to NCBI’s SRA under the BioProject accessions (PRJNA1015965, PRJNA455445, and PRJNA1015964, respectively). The reference genome of Sp. passalidarum NRRL Y-27907T is publicly available (Wohlbach et al., 2011). The genome assemblies and annotations of Sc. xylosifermentans, Sc. amazonensis, and Sc. coipomoensis are available from the JGI fungal genome portal MycoCosm (https:/mycocosm.jgi.doe.gov) and have been deposited at DDBJ/EMBL/GenBank under the BioProject accessions PRJNA1015837, PRJNA1015963, and PRJNA460959, respectively. RNA-Seq enrichment analysis code is available at https://github.com/kathbarros/goea.