Orthogonal chemical genomics approaches reveal genomic targets for increasing anaerobic chemical tolerance in Zymomonas mobilis
J.B. Eckmann et al. "Orthogonal chemical genomics approaches reveal genomic targets for increasing anaerobic chemical tolerance in Zymomonas mobilis" mSystems (2026) 11:e01001-25 [DOI:10.1128/msystems.01001-25]
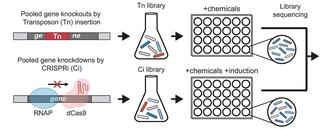
Genetically engineered microbes have the potential to increase efficiency in the bioeconomy by overcoming growth-limiting production stress. Screens of gene perturbation libraries against production stressors can identify high-value engineering targets, but follow-up experiments needed to guard against false positives are slow and resource-intensive. In principle, the use of orthogonal gene perturbation approaches could increase recovery of true positives over false positives because the strengths of one technique compensate for the weaknesses of the other, but, in practice, two parallel screens are rarely performed at the genome scale. Here, we screen genome-scale CRISPRi (CRISPR interference) knockdown and transposon insertion libraries of the bioenergy-relevant Alphaproteobacterium, Zymomonas mobilis, against growth inhibitors commonly found in deconstructed plant material. Integrating data from the two gene perturbation techniques, we established an approach for defining engineering targets with high specificity. This allowed us to identify all known genes in the cytochrome bc1 and cytochrome c synthesis pathway as potential targets for engineering resistance to phenolic acids under anaerobic conditions, a subset of which we validated using precise gene deletions. Strikingly, this finding is specific to the cytochrome bc1 and cytochrome c pathway and does not extend to other branches of the electron transport chain. We further show that exposure of Z. mobilis to ferulic acid causes substantial remodeling of the cell envelope proteome, as well as the downregulation of TonB-dependent transporters. Our work provides a generalizable strategy for identifying high-value engineering targets from gene perturbation screens that is broadly applicable.
Raw sequencing data for Tn and CRISPRi samples are available through the NCBI BioProjects PRJNA1270032 and PRJNA1276497, respectively. Raw proteomics data are available in the MassIVE database (MSV000097906). Code used for this work is available at https://github.com/GLBRC/orthogonal-chemgen-approaches-in-zmo.