Optimizing genomic prediction for complex traits via investigating multiple factors in switchgrass
P. Wang et al. "Optimizing genomic prediction for complex traits via investigating multiple factors in switchgrass" Plant Physiology (2025) 198:kiaf188 [DOI:10.1093/plphys/kiaf188]
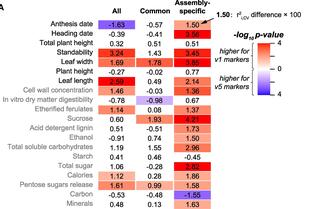
Genomic prediction has accelerated breeding processes and provided mechanistic insights into the genetic bases of complex traits. To further optimize genomic prediction, we assess the impact of genome assemblies, genotyping approaches, variant types, allelic complexities, polyploidy levels, and population structures on the prediction of 20 complex traits in switchgrass (Panicum virgatum L.), a perennial biofuel feedstock. Surprisingly, short read-based genome assembly performs comparably to or even better than long read-based assembly. Due to higher gene coverage, exome capture and multi-allelic variants outperform genotyping-by-sequencing and bi-allelic variants, respectively. Tetraploid models show higher prediction accuracy than octoploid models for most traits, likely due to the greater genetic distances among tetraploids. Depending on the trait in question, different types of variants need to be integrated for optimal predictions. Our study provides insights into the factors influencing genomic prediction outcomes, guiding best practices for future studies and for improving agronomic traits in switchgrass and other species through selective breeding.
All the scripts used in this study are available on GitHub at https://github.com/ShiuLab/Manuscript_Code/tree/master/2022_GP_in_Switchgrass.