Machine learning reveals genes impacting oxidative stress resistance across yeasts
K. Aranguiz et al. "Machine learning reveals genes impacting oxidative stress resistance across yeasts" Nature Communications (2025) 16:5866 [DOI:10.1038/s41467-025-60189-3]
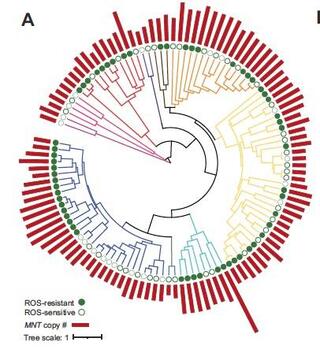
Reactive oxygen species (ROS) are highly reactive molecules encountered by yeasts during routine metabolism and during interactions with other organisms, including host infection. Here, we characterize the variation in resistance to the ROS-inducing compound tert-butyl hydroperoxide across the ancient yeast subphylum Saccharomycotina and use machine learning (ML) to identify gene families whose sizes are predictive of ROS resistance. The most predictive features are enriched in gene families related to cell wall organization and include two reductase gene families. We estimate the quantitative contributions of features to each species' classification to guide experimental validation and show that overexpression of the old yellow enzyme (OYE) reductase increases ROS resistance in Kluyveromyces lactis, while Saccharomyces cerevisiae mutants lacking multiple mannosyltransferase-encoding genes are hypersensitive to ROS. Altogether, this work provides a framework for how ML can uncover genetic mechanisms underlying trait variation across diverse species and inform trait manipulation for clinical and biotechnological applications.
The MS data generated for measuring protein levels in this study have been deposited in the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with the dataset identifier MSV000096915. Upon request from the corresponding author, all engineered strains are available under the Uniform Biological Material Transfer Agreement or another mutually agreeable material transfer agreement. Source data are provided with this paper. Scripts for building the ML model, model analysis, and generating figures associated with performance metrics are available on GitHub (https://github.com/katarinaaranguiz03/Yeast_ML_ROS)101. Versions of all dependencies and packages are also listed in the provided YAML file. A more general workshop and pipeline are also available on GitHub (https://github.com/ShiuLab/ML-Pipeline).