Directed Evolution Reveals Unexpected Epistatic Interactions That Alter Metabolic Regulation and Enable Anaerobic Xylose Use by Saccharomyces cerevisiae
T.K. Sato et al. (2016). Directed Evolution Reveals Unexpected Epistatic Interactions That Alter Metabolic Regulation and Enable Anaerobic Xylose Use by Saccharomyces cerevisiae. PLOS Genetics 12: e1006372
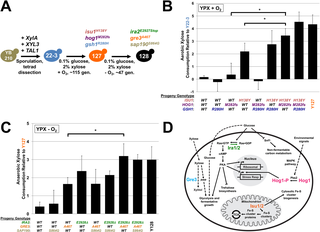
The inability of native Saccharomyces cerevisiae to convert xylose from plant biomass into biofuels remains a major challenge for the production of renewable bioenergy. Despite extensive knowledge of the regulatory networks controlling carbon metabolism in yeast, little is known about how to reprogram S. cerevisiae to ferment xylose at rates comparable to glucose. Here we combined genome sequencing, proteomic profiling, and metabolomic analyses to identify and characterize the responsible mutations in a series of evolved strains capable of metabolizing xylose aerobically or anaerobically. We report that rapid xylose conversion by engineered and evolved S. cerevisiae strains depends upon epistatic interactions among genes encoding a xylose reductase (GRE3), a component of MAP Kinase (MAPK) signaling (HOG1), a regulator of Protein Kinase A (PKA) signaling (IRA2), and a scaffolding protein for mitochondrial iron-sulfur (Fe-S) cluster biogenesis (ISU1). Interestingly, the mutation in IRA2only impacted anaerobic xylose consumption and required the loss of ISU1 function, indicating a previously unknown connection between PKA signaling, Fe-S cluster biogenesis, and anaerobiosis. Proteomic and metabolomic comparisons revealed that the xylose-metabolizing mutant strains exhibit altered metabolic pathways relative to the parental strain when grown in xylose. Further analyses revealed that interacting mutations in HOG1 and ISU1 unexpectedly elevated mitochondrial respiratory proteins and enabled rapid aerobic respiration of xylose and other non-fermentable carbon substrates. Our findings suggest a surprising connection between Fe-S cluster biogenesis and signaling that facilitates aerobic respiration and anaerobic fermentation of xylose, underscoring how much remains unknown about the eukaryotic signaling systems that regulate carbon metabolism.
All DNA sequencing reads have been deposited in the NCBI SRA under BioProject PRJNA279877. Raw data files for mass spectrometry proteomic data are available at https://chorusproject.org/pages/dashboard.html#/projects/all/1074/experiments (Project ID 1074).