Diagnosing and predicting mixed culture fermentations with unicellular and guild-based metabolic models
M.J. Scarborough et al. “Diagnosing and predicting mixed culture fermentations with unicellular and guild-based metabolic models” mSystems 5, 5 (2020) [DOI: 10.1128/mSystems.00755-20]
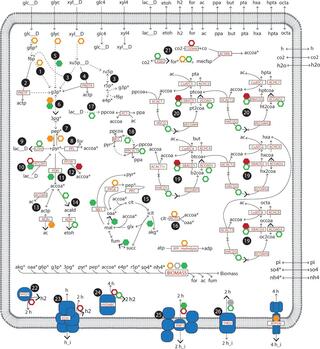
Microbiomes are vital to human health, agriculture, and protecting the environment. Predicting behavior of self-assembled or synthetic microbiomes, however, remains a challenge. In this work, we used unicellular and guild-based metabolic models to investigate the production of medium-chain fatty acids by a mixed microbial community that is fed multiple organic substrates. Modeling results provided insights into metabolic pathways of three medium-chain fatty acid-producing guilds and identified potential strategies to increase the production of medium-chain fatty acids. We constructed two metabolic models of MCFA-producing microbial communities based on available genomic, transcriptomic, and metabolomic data. This work demonstrates the role of metabolic models in augmenting multi-omic studies to gain greater insights into microbiome behavior.
First-generation predictive metabolic models of a mixed-culture fermentation can be found on GitHub.